Introduction
Methods and Materials
Plant Materials
Phenotypic Characteristic Analysis
Flow Cytometry Analysis
Root Collection and Chromosome Preparation
Fluorescence in Situ Hybridization (FISH)
Karyotype Analysis
Results and Discussion
Phenotypic Characteristics
Flow Cytometry Analysis
Chromosome Counting
Fluorescence in Situ Hybridization (FISH)
Karyomorphology Analysis
Conclusion
Introduction
Hibiscus belongs to the Malvaceae family and is a polymorphic genus of 250 ‑ 300 species of trees, shrubs, and herbs that grow throughout the world in tropical, subtropical, and temperate regions (Jo et al., 2019). H. sabdariffa is composed of tropical and subtropical shrubs and has many uses (Babajide et al., 2004; Mahadevan and Kamboj, 2009). Based on growth habit or use, H. sabdariffa is classified mainly under two varieties, H. sabdariffa var. altissima and H. sabdariffa var. sabdariffa (Mahadevan and Kamboj, 2009). H. sabdariffa var. sabdariffa is native to Asia (India to Malaysia), where it is also frequently cultivated (Mahadevan and Kamboj, 2009). The thick, fleshy, and cup-shaped calyces of H. sabdariffa var. sabdariffa are used to make jam, liquor, jelly, and tea, and are used to treat several human ailments (Faraji and Tarkhani, 1999; Chen et al., 2013; Da-Costa-Rocha et al., 2014). Tea made from H. sabdariffa var. sabdariffa is a popular beverage throughout the world (Clydesdale et al., 1979). Red calyces contain high amounts of ascorbic acid, riboflavin, niacin, calcium, and iron (Babalola et al., 2001; Wong et al., 2002; Fasoyiro et al., 2005; Qi et al., 2005). This plant is a short, bushy annual with green, red-streaked, and edible calyces, leaves, and seeds. In some countries, its flowers are used for decorative purposes (Sánchez-Mendoza et al., 2008).
Due to its huge economic value, making genetic improvements to H. sabdariffa var. sabdariffa is an important research area (Konar et al., 2018). Notwithstanding, cytogenetic studies are often hampered by the small size of mitotic chromosomes (Andras et al., 1999). Fluorescence in situ hybridization (FISH) and karyotyping are efficient techniques for cytogenetic studies (Anamthawat-Jónsson, 2004). FISH data can provide basic information on the ploidy level and chromosome characteristics of each species. Karyotyping is one of the prime cytological methods employed in empirical and theoretical research (Liang and Chen, 2015). Karyotype analysis of plants with a high number of chromosomes and small chromosomes is labor-intensive and difficult due to the differences in the physical features of the homologues and similarities of chromosomes in a complement (Silva et al., 2018). Moreover, genome size is a significant character of living organisms and estimates of genome size have been useful in systematic and evolutionary studies (Knight et al., 2005). However, molecular cytogenetic research of H. sabdariffa var. sabdariffa has rarely been reported. The flowers of this plant are cleistogamous, so conventional hybridization is difficult (Konar et al., 2018). Moreover, due to the fact that it is a tetraploid plant, the segregating populations need a longer period for segregation when a conventional genetic approach is used (Akpan, 2000). For this reason, molecular cytogenetic analysis is essential for accurate and efficient breeding. The objective of this study was to determine the genome sizes, DNA contents, cytogenetic characteristics, and morphological features of H. sabdariffa var. sabdariffa that will aid further research and breeding programs.
Methods and Materials
Plant Materials
Seeds of Hibiscus sabdariffa var. sabdariffa were collected from the Research Institute of Rose of Sharon and Tiger Lily, ‘Cheonan, Republic of Korea’. Seedlings were grown under greenhouse conditions at Kyungpook National University, Daegu, Republic of Korea. The seedlings were 1.5 years old when roots were collected.
Phenotypic Characteristic Analysis
Vegetative and reproductive characteristics were investigated followed by the survey method of the Korean Seed and Variety Service (htt://www.seed.go.kr). The characteristics of H. sabdariffa var. sabdariffa are represented in Table 1 and Fig. 1.
Table 1.
The phenotypic (vegetative and reproductive) characteristics of H. sabdariffa var. sabdariffa
Flow Cytometry Analysis
DNA content and genome sizes were measured by flow cytometry (Partec PA, Ploidy Analyzer, Sysmex, Kobe, Japan) with five randomly collected recently expanded leaves. Leaves were cut to approximately 1 cm2 and chopped in 500 µL of extraction buffer solution (Sysmex) in a petri dish and shaken for 30 s. The resulting solution was poured through a 30-µm nylon mesh filter into a 3-mL tube and 2 µL staining buffer was added. Finally, the nuclei suspension was injected into the flow cytometry analyzer.
Root Collection and Chromosome Preparation
Healthy root tips were collected and treated with an 8-hydroxyquinoline solution for 1 ‑ 2 h at room temperature. The root tips were then washed with double-distilled water and transferred into an aceto-ethanol (1:3, v/v) solution to fix the roots overnight. After rinsing with distilled water, 70% ethanol was used to preserve the roots at ‑ 20°C until further use. For slide preparation, the root tips were washed with distilled water to remove the ethanol solution and incubated with an enzyme mixture consisting of 0.3% pectolyase (Duchefa, Haarlem, The Netherlands), 0.3% cellulose (Duchefa), and 0.3% cytohelicase (Sigma, St. Louis, MO, USA) at 37°C for 40 min. Digested root tips were transferred to a clean slide and squashed under a microscope. Finally, 20 µL acetic acid (60%) was added per slide to spread the cells and the slides were air-dried.
Fluorescence in Situ Hybridization (FISH)
FISH was performed according to Lim et al. (2001) with minor modifications. Briefly, slides were pre-treated with 100 µg·mL-1 RNase A at 37°C for 40 min. After rinsing with 2X SSC and fixing with 4% paraformaldehyde for 10 min, the slides were rinsed again with 2X SSC and dehydrated in an ethanol series (70%, 90%, and 100%), followed by air-drying. The hybridization mixture contained formamide, 50% dextran sulfate, 20X SSC, 10% sodium dodecyl sulfate, herring sperm DNA, and rDNA probes. The mixture was placed in a water bath for DNA denaturation at 70°C for 5 min and then placed on ice for 15 min for fixation. Forty microliters of the mixture was added to each slide, and a cover slip was placed on the slide without bubbles. Slides were placed in a water bath for hybridization at 80°C for 5 min and then placed in a container with wet tissue paper and incubated in a humid chamber at 37°C for 16 h. Slides were then washed with 2X SSC buffer for 5 min followed by 0.1X SSC buffer for 30 min at 42°C with shaking and 2X SSC buffer for 5 min. Slides were submerged in 1X detection buffer for 5 min. Streptavidin CY3 (Invitrogen, Carlsbad, CA, USA) and anti-digoxigenin fluorescein (Roche, Basel, Switzerland) were used to detect the labeled chromosomes. Slides with cover-slips were placed in a humid chamber to incubate at 37°C for 1 h. Three jars containing 1X detection buffer were incubated in the dark in a water bath at 37°C for 5 min followed by dehydration an ethanol series (70%, 90% and 100%). Slides were then air-dried in the dark and counterstained with 4'6-diamidino-2-phenylindole (DAPI) and Vectashield at a 2:100 ratio (Vector Laboratories, Burlingame, CA, USA). Slides were examined with a model Nikon BX 61 fluorescence microscope (Tokyo, Japan). Probe signals were analyzed using ultraviolet excitation filters. Cytovision and imaging software were used to acquire images of the chromosomes with 5S rDNA and 18S rDNA signals.
Karyotype Analysis
For karyotype analysis, five cells showing well-spread metaphase chromosomes were selected. The length of each chromosome was measured by software, and chromosome number was determined based on the basis of short arm length order according to Lim et al. (2001). Chromosome types were classified according to the ratio of the short arm to the long arm (Levan et al., 1964). The image of the chromosomes was captured with 1000× magnification.
Results and Discussion
Phenotypic Characteristics
Leaf, flower, and calyx characteristics of H. sabdariffa var. sabdariffa are presented in Table 1 and Fig. 1. Leaf shape, apex, and base were ovate, acute, and rounded, respectively (Table 1), similar to Hibiscus hybrids ‘Tohagolred’ and ‘Daewangchun’(Jo et al., 2019). The length and width were 10.87 and 8.93 cm, respectively (Table 1), higher than those of two hibiscus hybrids ‘Tohagolred’(7.2 and 5.7) and ‘Daewangchun’ (8.4 and 5.4) (Jo et al., 2019). Moreover, flowers of H. sabdariffa var. sabdariffa were comparatively smaller than those of other Hibiscus species. Flower diameter, length, and width were 7.2, 3.07, and 2.77 cm, respectively, while ‘Daewangchun’ flowers were 16 cm in diameter (Kim et al., 2016). Fresh and dry weights of calyces, the most important part of H. sabdariffa var. sabdariffa, were 8.03 and 3.00 g, respectively.
Flow Cytometry Analysis
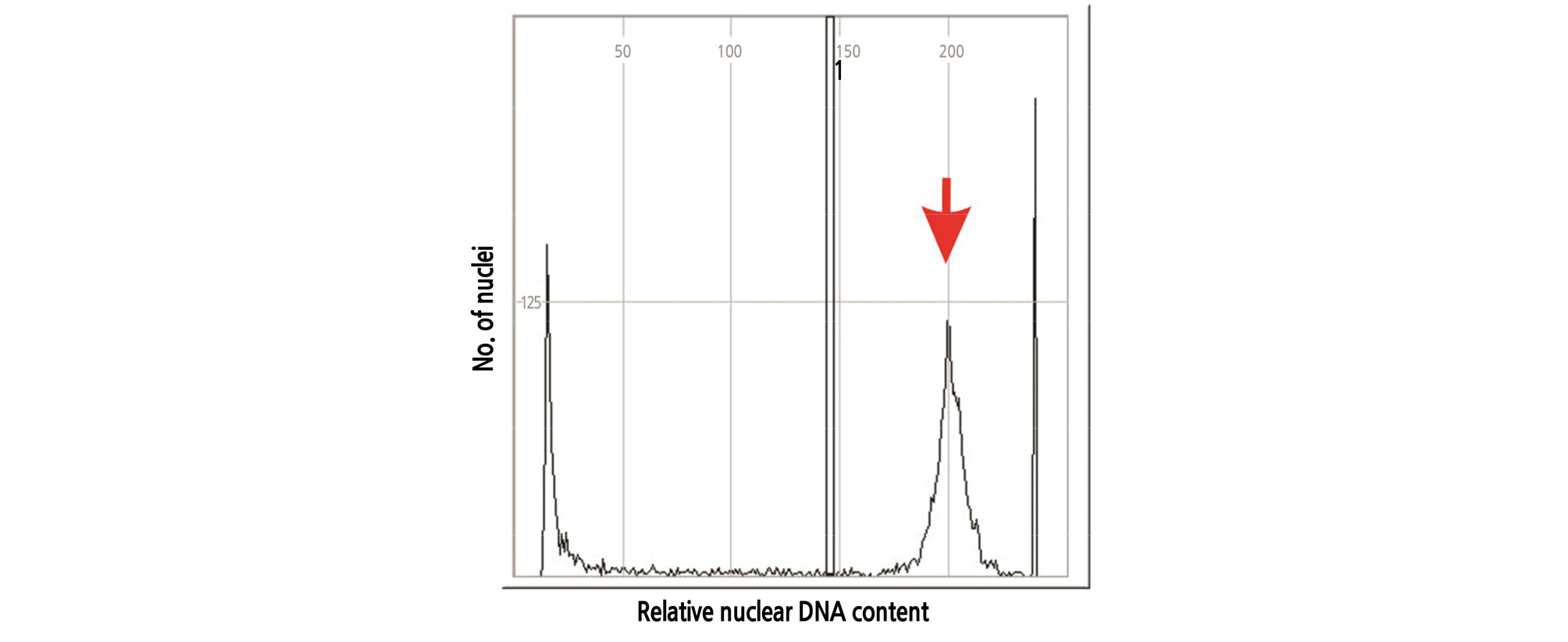
Flow cytometry is often used for hybrid identification and to measure nuclear DNA and genome sizes in plants (Dolezel et at., 2007). In this experiment, DNA content and genome sizes of H. sabdariffa var. sabdariffa were estimated by flow cytometry. According to our study, 2C-DNA content and 2C genome size were 4.29 pg and 3338.8 Mbp, respectively (Table 3 and Fig. 2), which was almost 32-fold larger than those of the angiosperm Genliea margaretae Hutch with the smallest genome size (Pellicer et at., 2010). Lattier at et. (2019) reported that the range of 2C-DNA values of various cytotypes of H. syriacus was from 4.55 to 8.98 pg and that H. hamabo (2C=4.06 pg)was also closer to H. syriacus.
Chromosome Counting
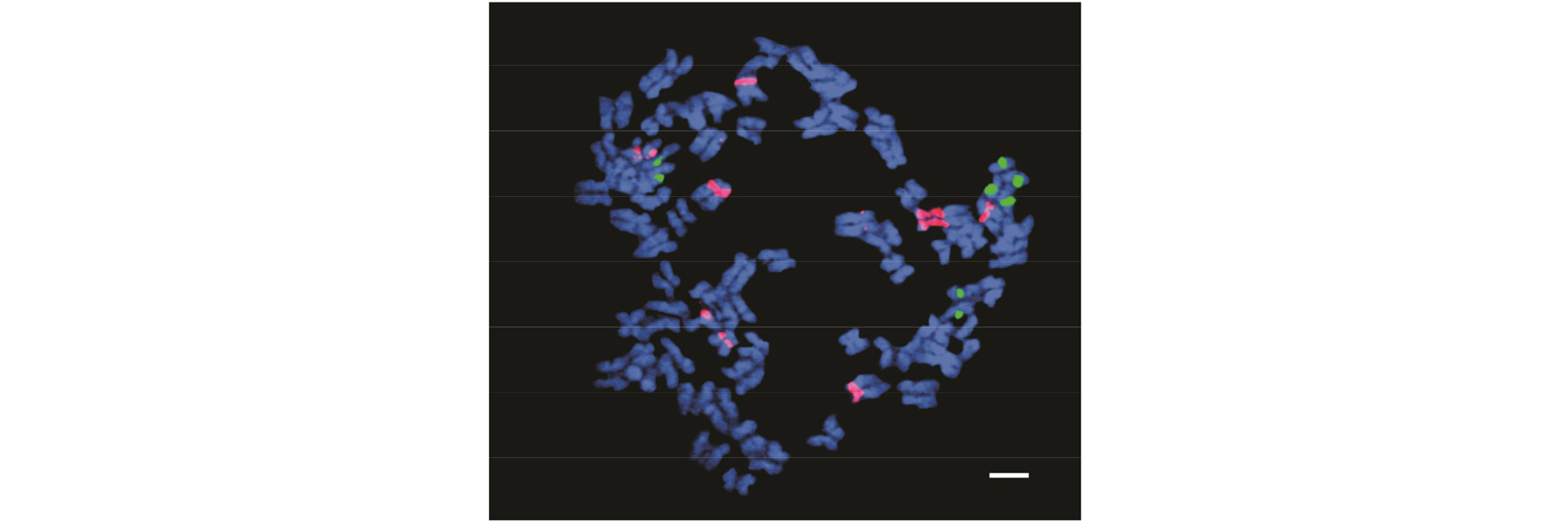
Detecting the chromosome number is a basic tool in cytogenetic analysis. After counting 100 cells, it was determined that the chromosome number of the somatic cells of H. sabdariffa var. sabdariffa was 72 (2n = 4x = 72) (Fig. 3). According to Akpan (2000), H. sabdariffa L was tetraploid (2n = 4x = 72) and the chromosome number was 72, which was consistent with the findings of this study. However, the chromosome number and ploidy levels of Hibiscuses were variable (Li et al., 2015b). Song (2001) reported the following: H. schizopetalus; 2n = 42, H. mutabilis; 2n = 92, H. rosa-sinensis; 2n = 4x = 84, H. rosa-sinensis,‘Double Rainbow’ 2n = 5x = 105, H. rosa-sinensis, ‘Flavo-plenus’ 2n = 6x = 138, and H. rosa-sinensis, ‘Carminatus’ 2n = 7x = 147. Therefore, the basic chromosome number of H. syriacus was x = 20 and most cultivars of H. syriacus and H. sinosyriacus were tetraploid, 2n = 4x = 80 (Skovsted, 1941; Lattier et al., 2019).
Fluorescence in Situ Hybridization (FISH)
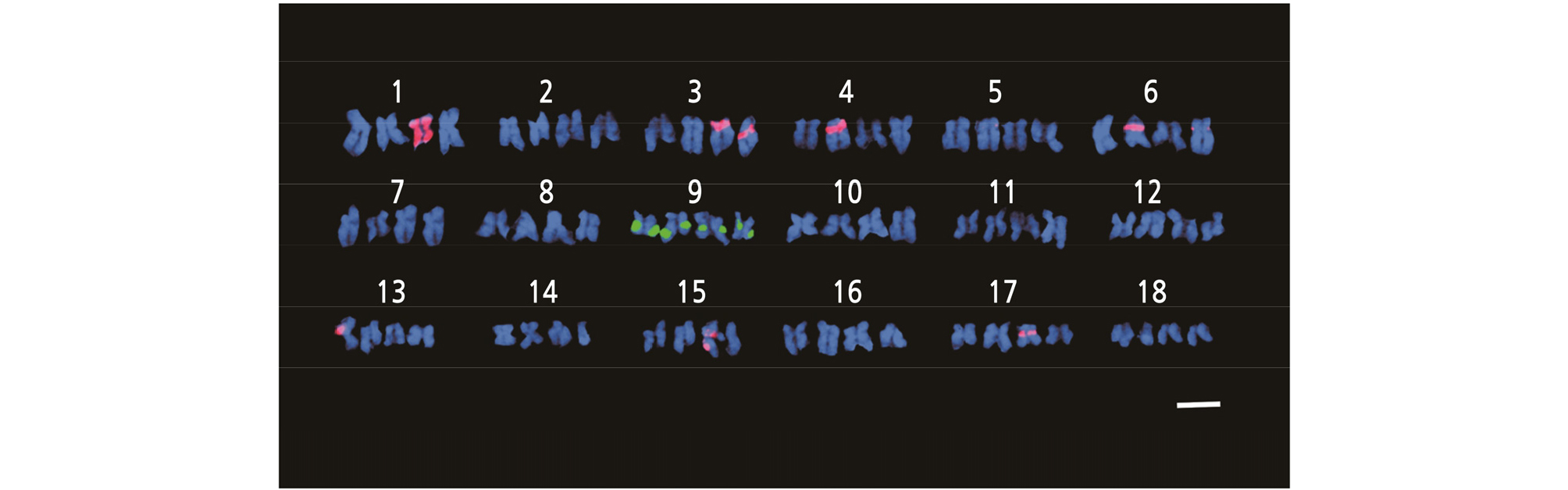
Chromosome analysis in H. sabdariffa var. sabdariffa by FISH is difficult and tedious due to the high number and small size of chromosomes. FISH is a molecular cytogenetic approach that detects complementary DNA sequences in nuclei using fluorescent-labeled probes (Rudkin and Stollar, 1977; Trask, 2002; Speicher and Carter, 2005; Hwang et al., 2011). It is a powerful tool to construct detailed karyotypes and observe chromosome morphology, such as centromere positions, arm lengths, and distribution of rDNA signals. Here, FISH using 5S rDNA and 18S rDNA probes was conducted. According to the FISH results, four 5S rDNA loci (green fluorescence) and ten 18S rDNA loci (red fluorescence) were observed on chromosome number 9 and chromosome numbers 1, 3, 4, 5, 6, 13, 15, and 17 (Figs. 4, 5 and Table 2). Among the ten red fluorescence signals (18S rDNA), eight were strong and two were weak. All four green fluorescence signals (5S rDNA) were strong (Figs. 4 and 5). Six red signals were located on the short arms of chromosome numbers 3, 4, 5, and 6. One red signal was found on the long arm of chromosome 15, and two signals were seen in the centromere region of chromosome numbers 13 and 17 (Fig. 5). The red signal on chromosome number 1 exhibited a special characteristic: the fluorescence extended along the entire chromosome except in the telometric region (Figs. 4 and 5). In terms of the green fluorescence signals (5S rDNA), all were observed on the long arm of chromosome number 9.
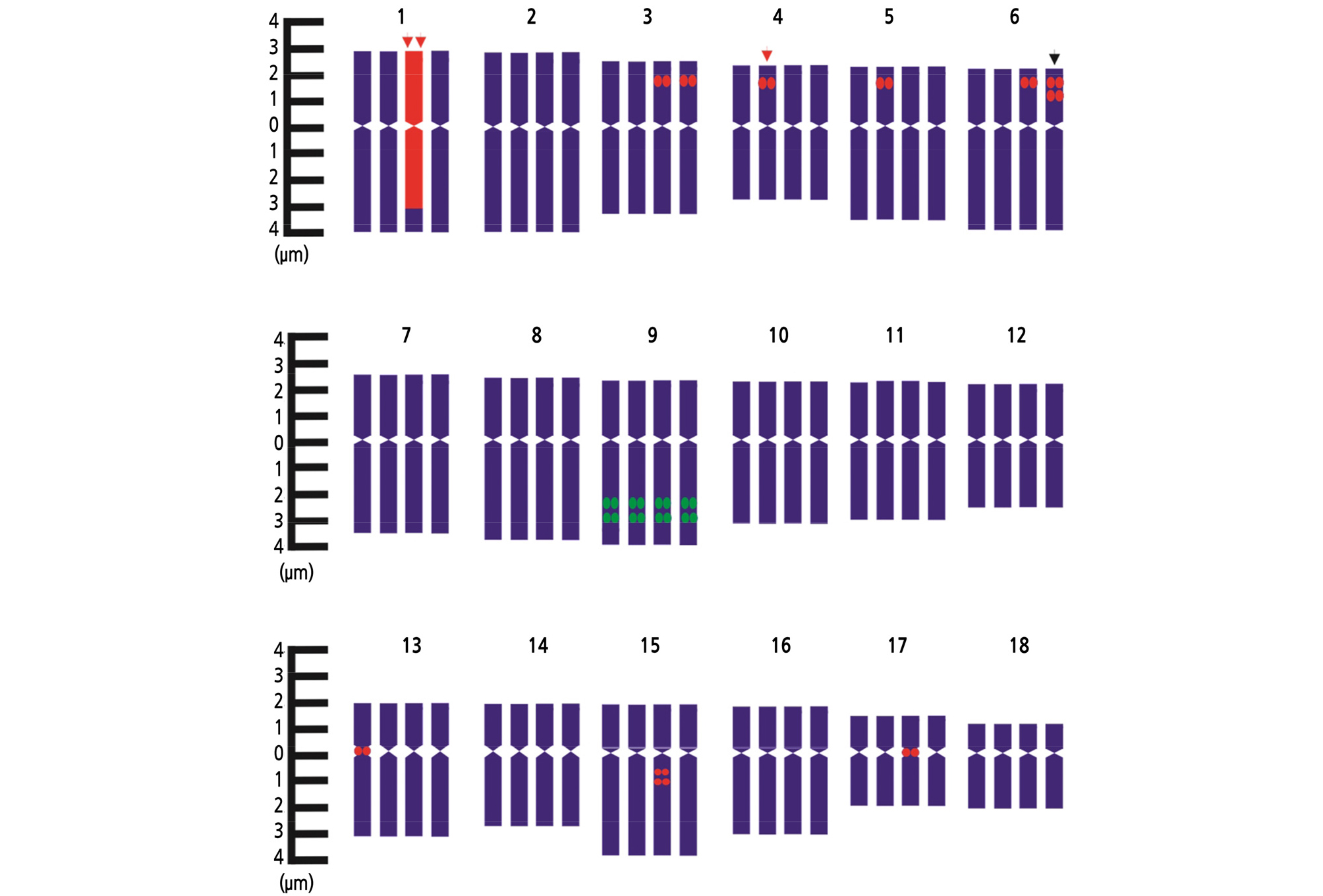
Fig. 5.
FISH karyotypic details of 5S rDNA and 18S rDNA loci on chromosomes of H. sabdariffa var. sabdariffa, where green fluorescence indicates 5S rDNA and red fluorescence indicates 18S rDNA. (Black arrow = weak signal, single red arrow = strong signals and double red arrow = special characteristic).
Table 2.
Chromosome length and distribution in FISH probes 5S and 18S rDNA of H. sabdariffa var. sabdariffa
Table 3.
Mean 2C (pg) nuclear DNA content and genome size (Mbp) of H. sabdariffa var. sabdariffa estimated by flow cytometry
| Variable | Mean ± SE | Minimum | Maximum | Median |
| 2C-DNA (pg) | 4.29 ± 0.25z | 4.26 | 4.32 | 4.29 |
| 2C genome size (Mbp) | 3338.8 ± 145.21z | 3038.38 | 3473.02 | 3337 |
Since Lim et al. (2000) reported the development of rDNA, cytogenetic research has advanced greatly. FISH has proven to be an efficient technique for cytological studies in woody angiosperms (Prado et al., 1996). Jo et al. (2019) used FISH to determine the number of 45S rDNA and 5S rDNA signals in H. syriacus ‘Samchully’ and found three 5S rDNA and six 45 rDNA loci. In this study, four 5S rDNA loci and ten 18S rDNA loci were detected. A study of cotton (Gossypium), a close relative to Hibiscus, produced the similar result. Most diploids had two 5S rDNA loci and all allotetraploid species had four rDNA loci (Gan et al., 2013).
Karyomorphology Analysis
The predominant chromosome size of Hibiscus was very small; in this study, chromosome length of H. sabdariffa var. sabdariffa ranged from 3.53 to 6.88 µm (Table 2), which was slightly longer than those of the chromosomes of H. paramutabilis (1.4 ‑ 4.8 µm) (Van Laere et al., 2010) and shorter than those of the diploid Lilium triginum (15.92 ‑ 30.18 µm) (Hwang et al., 2011). The longest chromosome was number 1, and the shortest chromosome was number 18 (Table 2 and Fig. 5).
The total chromosome length was 97.95 µm, and the total lengths of short arms and long arms were 37.16 and 60.79 µm, respectively (Table 2). The chromosome complement was composed of twelve pairs of metacentric chromosomes (1, 2, 3, 4, 5, 7, 8, 10, 12, 14, 17, and 18) and six pairs of submetacentric chromosomes (6, 9, 11, 13 15, and 16). No telocentric or subtelocentric chromosomes were observed (Table 2 and Figs. 4, 5). Therefore, the karyotype formula was K = (2n) = 4x = 72 = 48m + 24sm. Typically, chromosomes of Lilium were familiar as a complement composed of two large metacentric and ten subtelocentric chromosomes (Stewart, 1947). Hwang et al. (2011) reported two pairs of metacentric, four pairs of submetacentric, and six pairs of telocentric chromosomes in diploid L. trigrinum and two pairs of metacentric, six pairs of subtelocentric, and four pairs of telocentric in triploid L. trigrinum. However, in this study, twelve pairs of metacentric and six pairs of submetacentric chromosomes were observed. Different species have different karyotypes, not only between plant families, subfamilies, genera, and species, but also in different populations of the same species (Shi et al., 2009). Karyotype formulas of H. mutabilis and H. syriacus are K = (2n) = 2x = 92 = 86m + 6sm, and K (2n) = 4x = 56 = 32m + 16sm + 8st, respectively (Li et al., 2015a). Karyotype analysis is used to study and provide a basis for biological system development and relative relationships (Tabassum et al., 2014). Compared with simple shape classification, chromosome and karyotype data address the difficulties of interpreting conventional shape classification (Hu et al., 2002; Yan et al., 2008; Yang et al., 2008; Zhu and Wang, 2008).
Conclusion
This paper reported the phonotypic and cytogenetic analysis of H. sabdariffa var. sabdariffa. The phonotypic and cytogenetic characteristics such as the chromosome number, chromosome morphology, chromosome distribution of 5S and 18S rDNA, location of 5S and 18S rDNA signals on short and long arms, and karyotype, have been presented. Chromosome 1 exhibited 18S probe signals along the entire chromosome except the telometric region; this was a special characteristic and deserves further research. This study will help to provide a base for further research and contribute to the breeding of H. sabdariffa var. sabdariffa.