Introduction
Materials and Methods
Plant Materials and DNA Isolation
SNP Markers and Fluidigm Genotyping
Genetic Classification
Results
SNP Marker Analysis
Genetic Purity in F1 Hybrid Plants
Genetic Relationship Analysis
Discussion
Introduction
Cucumis melo is an important vegetable crop and includes several subspecies, where introgression from unknown germplasms continues to be a critical problem in breeding programs. In recent years, melon F1 hybrids were developed and released into the seed market by various seed companies (Nguyen et al., 2019; Kishor et al., 2020). The success of any breeding program mostly depends on an adequate supply of genetically pure hybrid seeds in the seed market. Therefore, seed companies require genetic purity tests, which are verified based on the success rate of cross-pollination and the number of plants with self-pollination, assuring good quality seeds.
In the past, genetic purity testing was performed by the grow-out test (GOT), which involves characterizing representative samples of F1 hybrid seeds into true hybrid seeds or off-types based on several morphological characteristics at various stages of plant growth (Pattanaik et al., 2018). However, this method is associated with many limitations. It is time-consuming, space-demanding, and yields ambiguous classification of the genotypes. Besides that, environmental influences on morphological characteristics also make it difficult to obtain accurate morphological data. In addition to the GOT, various biochemical methods, such as isozyme analysis followed by electrophoresis, have been used for the genetic purity testing of F1 hybrids (Markova et al., 2003; Jadhav and Achar, 2016). These techniques have advantages but are associated with limited polymorphism and environmental sensitivity.
Molecular markers are considered to be an efficient tool for genetic purity analysis due to their simple, fast, and accurate applications (Cheng-Xiang et al., 2005; Luan et al., 2010; An et al., 2010; Bae et al., 2015). Simple sequence repeats (SSRs) have become the most preferred marker for genetic purity analysis in melons (Cheng-Xiang et al., 2005; Luan et al., 2010). However, regions flanking SSRs could contain insertions or deletions (INDELs) or other SSR events (Bang and Chung, 2015) and the origin of the total length variation of the SSR markers should be confirmed by sequencing before F1 genetic purity analysis. Therefore, employing single nucleotide polymorphism (SNP)-based markers for F1 genetic purity analysis can overcome the limitations of SSR markers. SNPs are the most ideal markers because of their high abundance, even distribution, and strong marker-trait associations (Hayward et al., 2012), thus capturing importance for genetic diversity studies (Heo et al., 2017; Li et al., 2019).
The present study assessed the genetic purity of hybrid seeds of various commercial melons using genome-wide SNP markers developed by our group (Kishor et al., 2020). This study aimed to validate these genome-wide SNP markers to assess the genetic purity of F1sand PT breeding lines in melons.
Materials and Methods
Plant Materials and DNA Isolation
Eight PT breeding lines, 7_PT1, 10_PT10, 35-1_PT35, 46-1_PT46, 2H104_PT104, 2H106_PT106, PT1_male, and PT1_female, and 85 F1 hybrid plants derived from the cross between PT1_male and PT1_female were used in this study. All the eight PT melon breeding lines and 85 F1 hybrid plants were developed at Changchun Jongmyo Co., Ltd., Chilkog, Republic of Korea. The young leaves of 85 F1 hybrid plants, their parents, and other PT melon breeding lines were obtained and subjected to DNA extraction. Total genomic DNA (gDNA) was isolated from leaf tissue using the SDS procedure with slight modifications (Kim et al., 1997). The quality and quantity of DNA were determined by measuring the O.D. at 260/280 nm using a DS-11 spectrophotometer (Denovix Inc., DE, USA) followed by 1.2% gel electrophoresis.
SNP Markers and Fluidigm Genotyping
A total of 96 genome-wide SNP markers were obtained from our recent study (Kishor et al., 2020) and used for the genetic purity analysis of 85 F1 hybrid melon plants and their parents (Table 1).
Table 1.
The selected 96 SNPs for genetic purity analysis
Genotyping was conducted using the Fluidigm Juno system (Fluidigm Corporation, CA, USA). The step involving pre-amplification was performed using both specific-target amplification (STA) and locus-specific primers (LSP), followed by dilution of the pre-amplified products with distilled water. PCR amplification was performed using a set of allele-specific primers (ASP). End-point reads were detected using a Biomark EP1Reader (Fluidigm Corporation, CA, USA).The SNP calling was performed according to the Fluidigm Juno protocol using the Fluidigm SNP Genotyping Analysis software.
Genetic Classification
Contamination of the 85 F1 hybrid plants with the genetic material of other varieties or species was determined using polymorphic SNP markers in the STRUCTURE 2.3.4 program (Falush et al., 2003). The burn-in period was performed using 100,000 iterations, followed by 100,000 Markov chain Monte Carlo (MCMC) iterations per run. The number of genetically distinct populations (K) was adjusted from 1 to 10, and the model was repeated three times for each K. The best K value was estimated based on the delta K (ΔK) value using STRUCTURE HARVESTER (Earl and von Holdt, 2012). Similarly, an unweighted pair group method with arithmetic average (UPGMA) tree was constructed using Cavalli-Sforza and Edwards’ (1967) genetic distance method in PowerMarker V3.25 (Liu and Muse, 2005). The UPGMA tree was constructed using the SNP assay results of 95 commercial melon cultivars (M1 to M95) from our previous study (Kishor et al., 2020), and the SNP assay results of the present study.
Results
SNP Marker Analysis
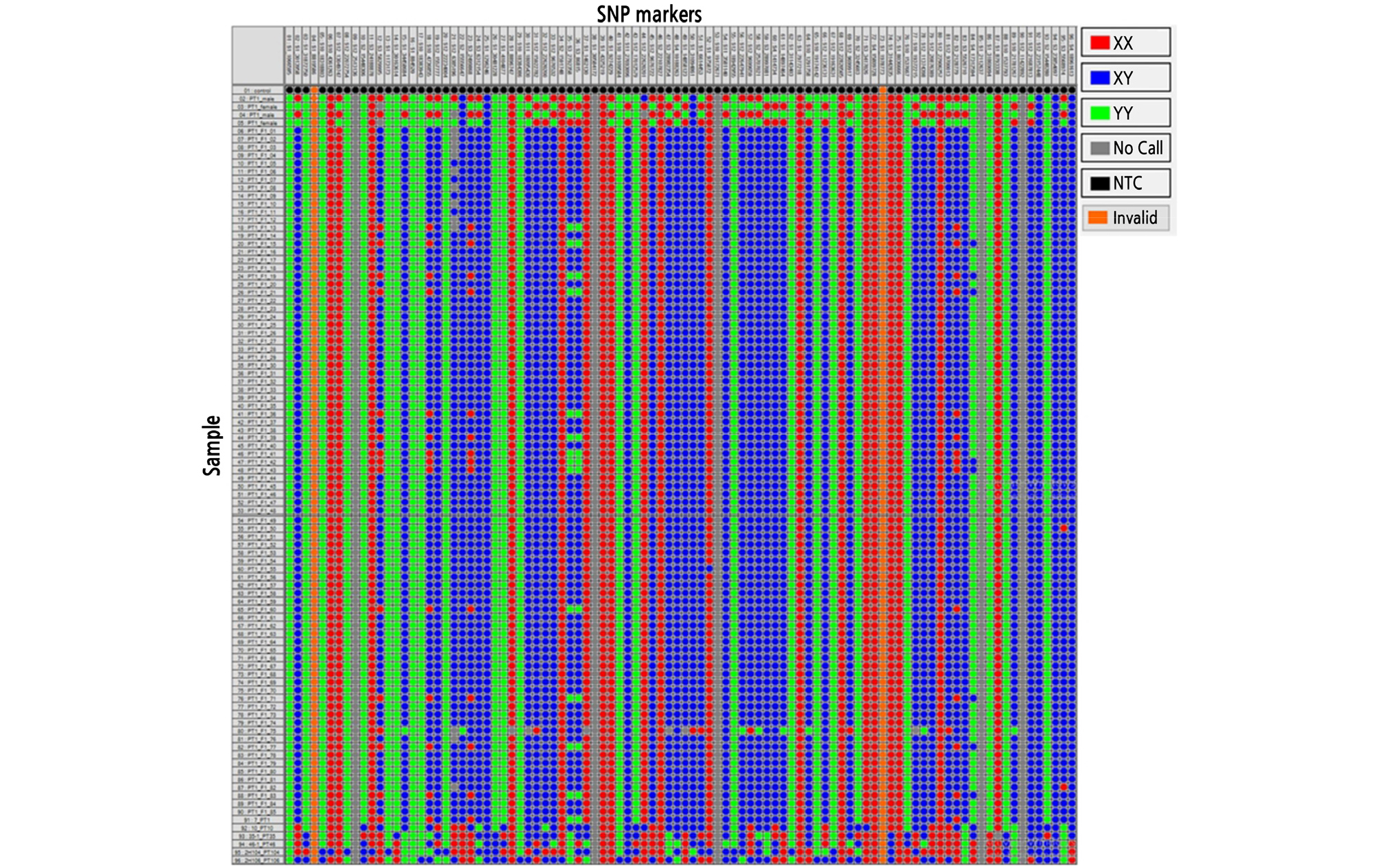
From the 96 SNP marker assay in the 85 F1 hybrid plants, their parents, and other PT melon breeding lines (Fig. 1), 89 SNP markers (92.70%) had successful DNA amplification, of which 39 SNP (43.82%) markers showed stable polymorphisms between PT1_male and PT1_female (Table 1). Therefore, these 39 polymorphic SNP markers were used to distinguish the F1 hybrid plants and their parents in the present study (Table 1). In contrast, the other 50 SNP markers could not amplify the DNA in Fluidigm genotyping but displayed no calls, monomorphism, or heterozygosity in the F1 hybrid plants and their parents.
Based on the six markers, M3_5822995, M10_4736855, M3_2738358, M3_86815, M3_2128779, and M4_17312594, F1 hybrid plant numbers PT1 F1 13, PT1 F1 15, PT1 F1 19, PT1 F1 21, PT1 F1 36, PT1 F1 39, PT1 F1 41, PT1 F1 42, PT1 F1 43, PT1 F1 60, PT1 F1 71, PT1 F1 77, and PT1 F1 83 showed amplification errors in the SNP assay. Additionally, F1 hybrid plant numbers PT1 F1 50 and PT1 F1 82 showed amplification errors in the M3_5568974 SNP marker only. These results suggest that amplification errors might be associated with outcrossing. Similarly, a genotyping error was also observed in F1 hybrid plant number PT1 F1 55 for the M1_975872 monomorphic SNP marker, which could be due to the amplification of a non-target site. In contrast, all 89 SNP markers successfully discriminated genotypes such as 7_PT1, 10_PT10, 35-1_PT35, 46-1_PT46, 2H104_PT104, and 2H106_PT106 in the SNP assay.
Genetic Purity in F1 Hybrid Plants
Contamination with other genetic material of other varieties or species was determined by using 39 polymorphic SNP markers in the model-based STRUCTURE program; this assumes many populations among 85 PT1 F1 hybrid plants. The delta K value was maximum at K=2 (Fig. S1). The individuals under the different populations with scores of more than 0.80 were classified as pure and scores of less than 0.80 as admixture plants. The results showed that F1 hybrid plant numbers PT1 F1 13, PT1 F1 15, PT1 F1 19, PT1 F1 21, PT1 F1 36, PT1 F1 39, PT1 F1 41, PT1 F1 42, PT1 F1 43, PT1 F1 60, PT1 F1 71, PT1 F1 77, and PT1 F1 83 were grouped into a separate cluster (Fig. 2), suggesting the contamination with other genetic material of other varieties due to outcrossing. Additionally, F1 hybrid plant number PT1 F1 75 showed admixed genetic material. Hence, these plants should not be considered for the selection process due to outcrossing.
Genetic Relationship Analysis
A UPGMA tree was constructed based on the SNP assay results of 96 SNP markers in 95 commercial melon cultivars (Kishor et al., 2020), and SNP assay results of the present study, which included 7_PT1, 10_PT10, 35-1_PT35, 46-1_PT46, 2H104_PT104, 2H106_PT106, PT1_males, and PT1_females, F1 hybrid plant numbers PT1 F1 01, PT1 F1 02, PT1 F1 13, PT1 F1 15, PT1 F1 19, PT1 F1 21, PT1 F1 36, PT1 F1 39, PT1 F1 41, PT1 F1 42, PT1 F1 43, PT1 F1 60, PT1 F1 71, PT1 F1 77, and PT1 F1 75, classified into eight distinct groups (Fig. 3). Most of the melon cultivars were distributed in group VIII, followed by group IV and group VII based on the 96 SNP markers. However, M37 deviated as a separate group I.
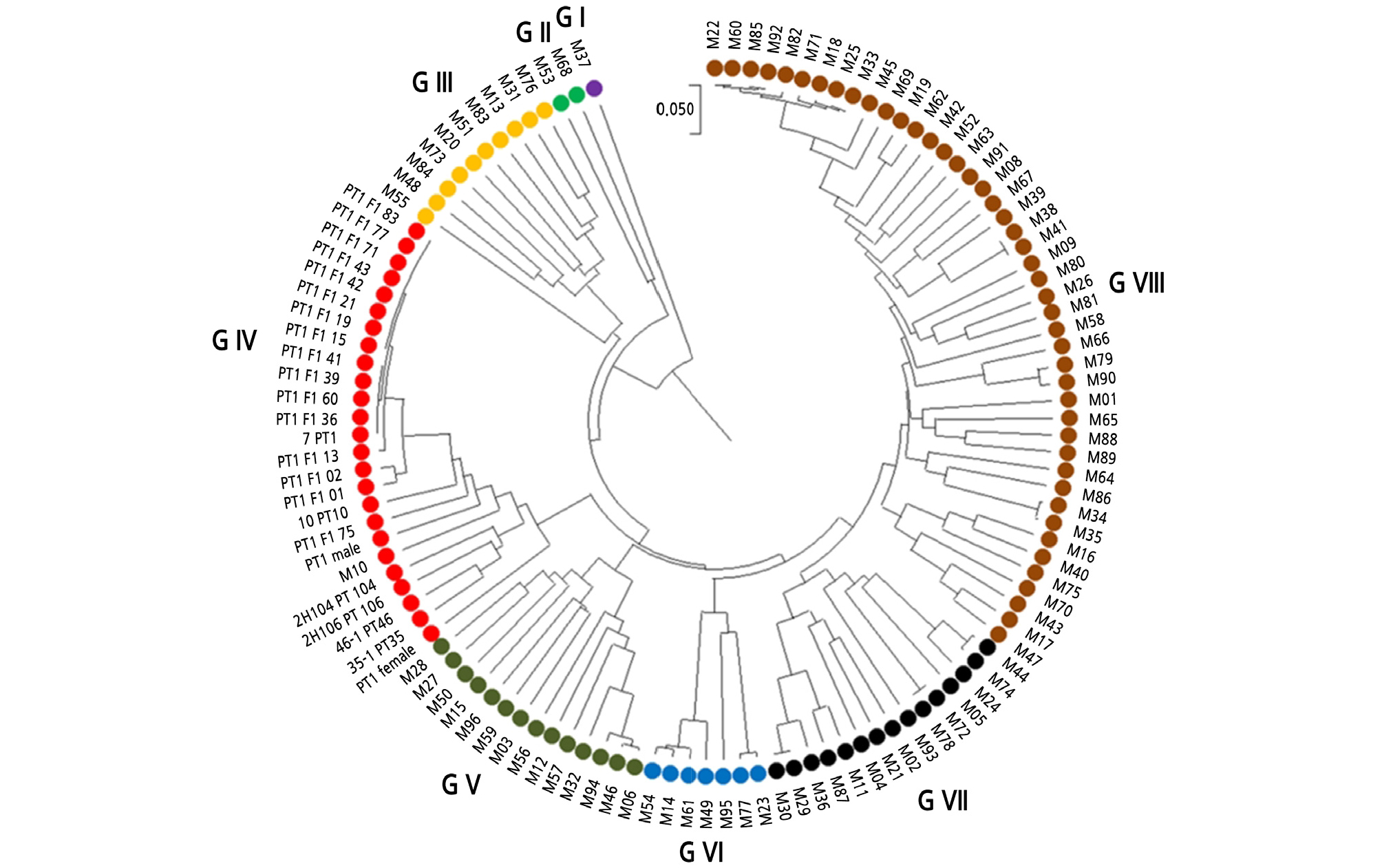
Fig. 3.
UPGMA dendrogram based on Cavalli-Sforza and Edwards’ (1967) genetic distance using 96 SNP markers in 119 samples consisting of 95 melon cultivars (M1 to M95) from our previous study (Kishor et al., 2020), PT1 F1 hybrid plants, and PT melon breeding lines. All eight distinct groups were assigned different colors.
All the PT1 melon breeding lines and F1 hybrid plant numbers were grouped with M10 in group IV. Interestingly, all the PT1 melon breeding lines, F1 hybrid plants, and M10 were developed at the Changchun Jongmyo Co., Ltd, Company, Chilkog, Republic of Korea. Additionally, F1 hybrid plant numbers PT1 F1 13, PT1 F1 15, PT1 F1 19, PT1 F1 21, PT1 F1 36, PT1 F1 39, PT1 F1 41, PT1 F1 42, PT1 F1 43, PT1 F1 60, PT1 F1 71, and PT1 F1 77 were closely sub-grouped with the 7_PT1 melon breeding line in group IV. However, F1 hybrid plant number PT1 F1 75 deviated as a separate sub-group in group IV.
Discussion
The genetic purity of hybrids and cultivars is of great importance for the success of any breeding program. In the present study, 85 F1 hybrid plantsand eight PT melon breeding lines were evaluated using genome-wide SNP markers to assess genetic purity.
Among the various DNA-based markers, SSR markers are predominantly used for genetic purity analysis of melons (Cheng-Xiang et al., 2005; Luan et al., 2010). However, these markers are associated with INDELs or other SSR events within its flanking region (Bang and Chung, 2015). Recently, Next-generation Sequencing (NGS) technology gaining importance to generate large number of SNPs and used to develop SNP markers (Jung et al., 2020; Kishor et al., 2020). Similarly, previous studies were also reported handful of high-resolution melting (HRM)-based SNP markers for genetic purity analysis of F1 hybrids (An et al., 2010) and powdery mildew race 5-specific SNP markers in Cucumis melo (Howlader et al., 2020). Presently, however, there are very limited numbers of these SNP markers for melons. In the latest study, we reported genome-wide SNP markers via genotyping-by-sequencing (GBS) in melons (Kishor et al., 2020). This study revealed that numerous high-quality SNP markers were distributed across the 12 chromosomes in the melon genome.
Here, we used 96 high-quality SNP markers from our previous study (Kishor et al., 2020) and performed genetic purity analysis of F1 hybrids and PT breeding lines in melon via Fluidigm SNP assays. The results indicated a 92.70% success rate of DNA amplification in both the 85 F1 hybrid plantsand the eight PT melon breeding lines. Although a total of 96 SNP markers were screened, 39 SNP (43.82%) markers showed stable polymorphisms between the PT1_male and the PT1_female. Such a result was associated with highly similar genetic backgrounds between the parents, possibly due to the relatively narrow genetic base in the crop plants (Pattanaik et al., 2018).
In a breeding program, seeds can become admixed due to several reasons, such as outcrossing, pollen shedders, and physical mixtures (Pattanaik et al., 2018). Therefore, providing genetically pure hybrid seed is very important for commercially successful hybrids. A recent study reported that admixed cultivars could be classified using population structure analysis in melons (Kishor et al., 2020).
In the present study, we identified several F1 hybrid plants associated with contamination due to outcrossing based on SNP marker analysis and population structure analysis. Additionally, the SNP analysis and population structure analysis results identifying the genetic purity of F1 hybrid plants were consistent with each other. Hence, these contaminated plants are not recommended for further selection processes. Further, UPGMA analysis revealed that most of the contaminated plants were closely sub-grouped with 7_PT1, suggesting possible outcrossing with the 7_PT1 melon breeding line.
Together with DNA extraction, the Fluidigm-based SNP marker analysis presents a simple and effective approach for quality testing of melon hybrids and breeding lines. Additionally, SNP marker technology could identify genetic similarities and differences by comparing melon PT breeding lines with commercial or registered melon cultivars. Therefore, these SNP markers can help breeders protect the plant proprietary rights of new cultivars or hybrids through genetic purity testing of the melons.
Supplementary Material
Supplementary materials are available at Horticultural Science and Technology website (https://www.hst-j.org).
- HORT_20200062_Fig_1s.pdf
Delta K values assumed based on STRUCTURE analysis.