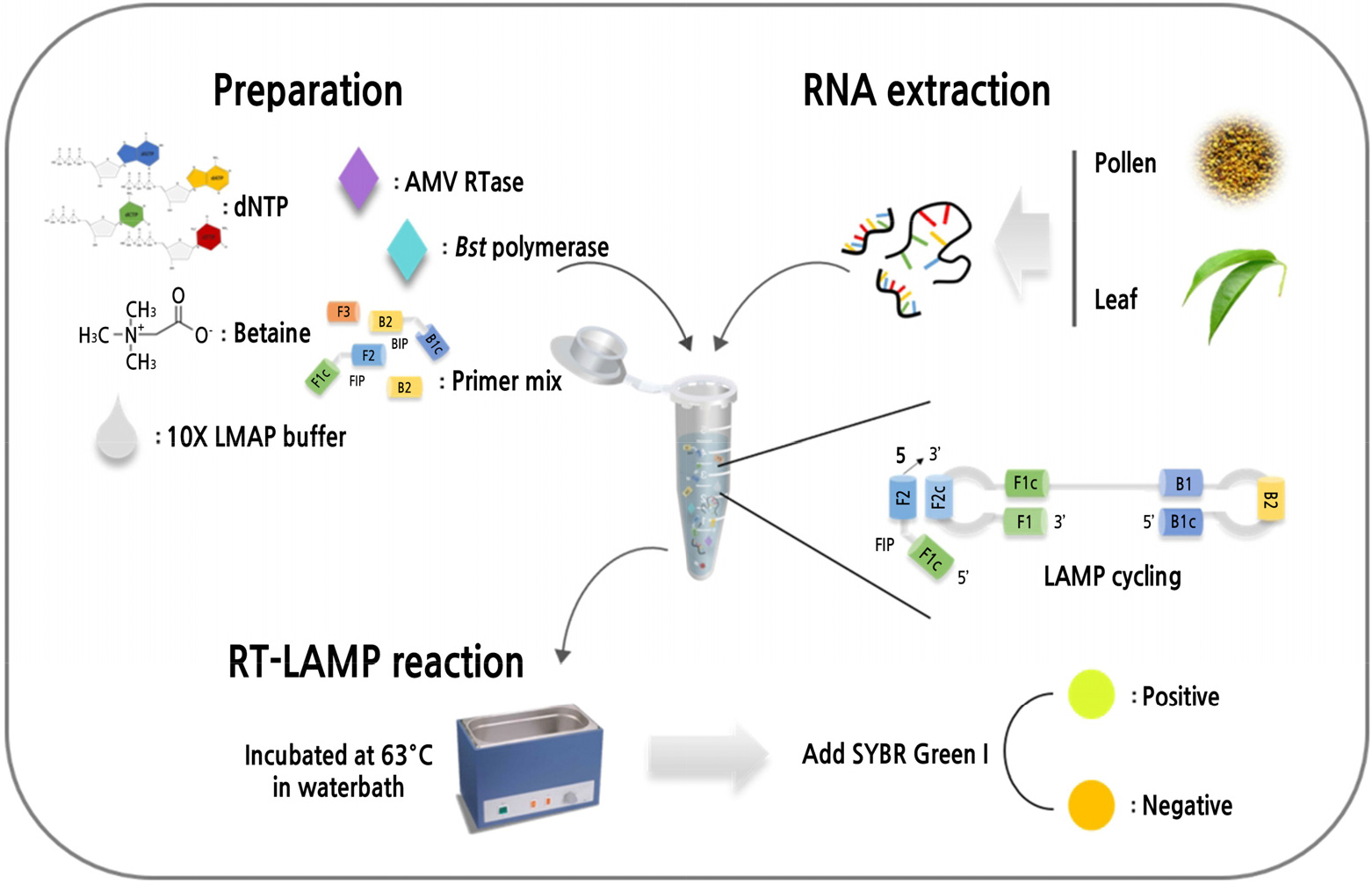
Introduction
Materials and Methods
Plant Materials
RNA Extraction
RT-PCR
RT-LAMP Primer Design
RT-LAMP Optimization
Specificity and Sensitivity of the RT-LAMP
Results
Primer Efficiency
RT-LAMP Optimization
Sensitivity and Specificity of RT-LAMP for PLMVd Detection
Detection of PLMVd in Field Samples
Discussion
Introduction
Following the international trend of integrating domestic and overseas markets, the Korean fruit industry has recently become much more globalized. However, simultaneous increases in fruit tree cultivation and reductions in pollinating insects have resulted in a sharp increase in artificial pollination, and over 90% of the pollen used for artificially pollinating apple, kiwi, peach, and pear is imported from other countries, including China, Australia, and the USA. Currently, more than 45 plant viruses (primarily Alphacryptovirus, Ilarvirus, Nepovirus, and Potyvirus species) and five viroids are reportedly transmissible through pollen transfer (Card et al., 2007; Liu et al., 2014). Most pollen-transmitted viruses have been reported to exhibit horizontal transmission, in which the virus is transmitted from the pollen to seeds through the infection of pollinated ovules. However, little is known about this process, and some pollen-transmitted viruses are transmitted vertically, in which maternal tissues are infected during pollination (Mandahar, 1985; Mink, 1993; Card et al., 2007; Vijeth et al., 2018). Because of their unique modes of transmission, these viruses can be stable in infected pollen and, as such, represent a significant risk to phytosanitation even after several years of storage (Shiller et al., 2010).
In particular, peach pollen has been reported to transmit prunus necrotic ringspot virus (PNRSV), cherry leaf roll virus (CLRV), prune dwarf virus (PDV), and cherry virus A (CVA; Beaver-Kanuya and Harper, 2019), as well as peach latent mosaic viroid (PLMVd), which belongs to the genus Pelamoviroid in the Avsunviroidae family of chloroplastic viroids with hammerhead ribozymes(Flores et al., 2006; Di Serio et al., 2018). The PLMVd genome is a 336- to 351-nucleotide circular RNA, depending on the inclusion of certain introns, and possesses a branched conformation structure that is stabilized by a pseudoknot between two kissing loops (Flores et al., 2006). To date, PLMVd has been reported to infect a variety of Prunus species, including apricot, cherry, pear, nectarine, plum, peach, and several peach hybrids (Dubé et al., 2009), and to induce mosaic and white patterns (peach calico) and chlorotic mosaic patterns in infected leaves, fruits, and flowers (Flores et al., 1990).
Currently, several molecular methods (e.g., RT-PCR and qRT-PCR) are used to detect viruses and viroids in imported pollen. However, PCR amplification requires the use of a thermal cycler to control the necessary temperature changes, and TaqMan-probed qRT-PCR assays are both slow (~100 min) and require expensive and specific equipment. As an alternative, several studies have reported the development of rapid and specific virus-detecting assays, such as reverse transcription loop-mediated isothermal amplification (RT-LAMP; Varga and James 2006; Johnson et al., 2014; Shen et al., 2014; Zhao et al., 2014; Zong et al., 2014; Almasi 2015; Peng et al., 2017; Tahzima et al., 2019). Indeed, RT-LAMP assays are highly sensitive and specific isothermal reactions that overcome the disadvantages of other amplification methods and possess a variety of advantages, including short amplification times and no requirement for expensive or specialized equipment (Notomi et al., 2000). Accordingly, the aim of the present study was to use imported peach pollen to develop an RT-LAMP assay for detecting PLMVd in quarantine sites. The application of an isothermal amplification assay is a challenge to the progress of the phytosanitary frameworks in imported pollen.
Materials and Methods
Plant Materials
Peach pollen that was imported from China for artificial pollination in 2018 and 2019 was purchased from commercial markets,and peach leaves that were infected with PLMVd, apple chlorotic leaf spot virus (ACLSV), and prunus necrotic ringspot virus (PNRSV) were provided kindly by the Animal and Plant Quarantine Agency of Korea. Pollen samples were pooled, ground in liquid nitrogen, and stored at ‑ 80°C.
RNA Extraction
Total RNA was extracted from peach pollen using the IQeasy Plus Plant RNA Extraction Mini Kit (iNtRON, Korea) according to the manufacturer’s protocol. Both the quantity and quality of the purified RNA samples were measured and evaluated using a Biochrom Libra S70 double-beam spectrophotometer (Biochrom, Ltd., England), and the RNA quality was further analyzed using electrophoresis on 1.2% (w/v) agarose gel in 0.5× TBE buffer with RedSafe nucleic acid staining (iNtRON, Daejeon, Korea). All the RNA samples were stored at ‑ 20°C.
RT-PCR
RT-PCR was performed using the 2× RT-PCR premix (SuPrimeScript; GeNet Bio, Korea) according to the manufacturer’s protocol. Each of the 20-µl reactions included 0.4 µM forward primer, 0.4 µM reverse primer, 10 µl 2× SuPrimeScript RT Premix, 2 µl template, and 6 µl DEPC-treated water, and amplification was performed under the following conditions: a reverse transcription step of 30 min at 50°C; initial denaturation at 95°C for 5 min; 35 cycles of 95°C for 30 sec, 56°C for 30 sec, and 72°C for 1 min; and a final extension of 5 min at 72°C. Previously published primer sets were used for the amplification of PLMVd (Animal and Plant Quarantine Agency of Korea), ACLSV (Bae, 2015), PNRSV (Kim et al., 2009), and nad5 (Menzel et al., 2002).
RT-LAMP Primer Design
The genome sequences of 27 PLMVd isolates were downloaded from NCBI (GenBank accessions: KY355293, KY355377, KX430156, GQ499315, JX479199, KF868404, JF898826, KY355300, KY355296, KY355333, KY355234, JF898825, KX430166, KX430164, KX430160, GQ499312, KF867859, KT005802, KT033052, KT033051, KY355381, KY355341, KY355301, KY355261, KY355201, and AY685181), and a multiple sequence alignment was performed using ClustalW in BioEdit v. 7.0.5.3. Based on the sequences of conserved regions of the PLMVd genome (GenBank accession: LC469622), four oligonucleotide RT-LAMP primers were then designed using PrimersExplorer V5 (http://primerexplorer.jp/ lampv5e/index.html). To identify optimal primers, six sets of PLMVd-specific RT-LAMP primers were designed (Table 1). The primers were synthesized by Bionics Co., Ltd. (Korea).
Table 1.
Primers used for the RT-LAMP detection of peach latent mosaic viroid (PLMVd)
| Primer name | Primer type | Sequence 5'-3' | Genome positionz | Length |
| PLMVd 1 | F3 | GCTAAGCACGTCGCAATGA | 127-145 | 19 bp |
| B3 | TTGCAGGAACTCGTCAGTG | 300-318 | 19 bp | |
| FIP (F1c-F2) | CCAGGATCACACTCCCCTCTGATTTTCGTAAGGTGGGGCTTTTCC | 186-207, 146-164 | 45 bp | |
| BIP (B1c-B2) | TGGATTACGACGTCTACCCGGGTTTTAGCACAGACCTTTCACTCAG | 223-244, 275-294 | 46 bp | |
| PLMVd 2 | F3 | ATGACGTAAGGTGGGGCT | 142-159 | 18 bp |
| B3 | GCAGGAACTCGTCAGTGTG | 298-316 | 19 bp | |
| FIP (F1c-F2) | CAGTTTCTACGGCGGTACCAGGTTTTTTTCCCTTGAAGTTGAGCGG | 203-224, 160-179 | 46 bp | |
| BIP (B1c-B2) | TACGACGTCTACCCGGGATTCATTTTAGCACAGACCTTTCACTCAG | 228-249, 275-294 | 46 bp | |
| PLMVd 3 | F3 | TGCCCTATCCGAGCACTG | 93-110 | 18 bp |
| B3 | TTGCAGGAACTCGTCAGTG | 300-318 | 19 bp | |
| FIP (F1c-F2) | TCCCCTCTGAAGTCGACCGCTTTTGCTAAGCACGTCGCAATGA | 176-195, 127-145 | 43 bp | |
| BIP (B1c-B2) | TGGATTACGACGTCTACCCGGGTTTTAGCACAGACCTTTCACTCAG | 223-244, 275-294 | 46 bp | |
| PLMVd 4 | F3 | TGCCCTATCCGAGCACTG | 93-110 | 18 bp |
| B3 | TTGCAGGAACTCGTCAGTG | 300-318 | 19 bp | |
| FIP (F1c-F2) | CGACCGCTCAACTTCAAGGGAATTTTCGGTAGAAAGGCTAAGCACG | 161-182, 117-136 | 46 bp | |
| BIP (B1c-B2) | TGGATTACGACGTCTACCCGGGTTTTAGCACAGACCTTTCACTCAG | 223-244, 275-294 | 46 bp | |
| PLMVd 5 | F3 | TGCTGCTTCGGTAGAAAGG | 109-127 | 19 bp |
| B3 | CGTTCCATTGCAGGAACTCG | 306-325 | 20 bp | |
| FIP (F1c-F2) | CCAGGATCACACTCCCCTCTGATTTTCGTAAGGTGGGGCTTTTCC | 186-207, 146-164 | 45 bp | |
| BIP (B1c-B2) | TACGACGTCTACCCGGGATTCATTTTCAGTGTGCTTAGCACAGACC | 228-249 | 46 bp | |
| PLMVd 6 | F3 | AGTTTCGTCGCATCTCAGC | 6-24 | 19 bp |
| B3 | CCAGGATCACACTCCCCT | 190-207 | 18 bp | |
| FIP (F1c-F2) | TAGGGCACCCCAAGGTGGAGTTTTACTCATCAGTGGGCTTTGC | 80-99, 26-44 | 43 bp | |
| BIP (B2-B1c) | AGAAAGGCTAAGCACGTCGCATTTTGAAGTCGACCGCTCAACTT | 121-141, 169-187 | 44 bp |
RT-LAMP Optimization
One-step RT-LAMP was performed using 25-µl reaction mixtures that each contained outer primers (PLMVd F3/B3, 0.4 µM), inner primers (PLMVd FIP/BIP, 1.6 µM), 2.5 µl 10× LAMP buffer [20 mM Tris-HCI (pH 8.8), 10 mM KCL, 10 mM (NH4)2SO4, 6 mM MgSO4, Tween 20%], 1 µl Bst DNA polymerase (Large fragment, 8 U/µl; Enzynomics, Korea), 0.5 µl AMV reverse transcriptase (30 U/µl; Promega, USA), 2.4 mM dNTPs (iNtRON), 0.8 M Betaine (≥98%, perchloric acid titration; Sigma-Aldrich, USA), 2 µl RNA template, and DEPC-treated water. Either water or RNA from PLMVd-free samples was used as a negative control. To identify the optimal reaction temperature, the RT-LAMP reactions were conducted at 54°C, 57°C, 60°C, 63°C, 66°C, 69°C, 72°C, or 75°C for 60 min in a water bath, followed by 5 min at 80°C in a thermocycler (Applied Biosystems, LifeScience Technologies, CA, USA), and to identify the optimum reaction time, the RT-LAMP reactions were conducted at 63°C for 5, 10, 20, 30, 40, 50, 60, or 70 min, followed by 5 min at 80°C. To assess amplification, 10 µl 100× SYBR Green I (diluted 1:100 in DEPC-treated water; Invitrogen, USA) was added to each reaction tube, and amplification success was assessed by direct visual inspection. The RT-LAMP amplification was also confirmed by visualizing 5 µl RedSafe stain on 2% (w/v) agarose gels using electrophoresis in 0.5× TBE buffer. The optimal temperature (63°C) and reaction time (60 min) were used for further experiments.
Specificity and Sensitivity of the RT-LAMP
The specificity of the RT-LAMP primers for PLMVd was examined using total RNA extracted from peach leaves infected with either ACLSV or PNRSV and with RNA from healthy peach leaves as a negative control. Meanwhile, the sensitivity of the optimized reaction conditions was assessed using 10-fold serial dilutions (100 to 10-7) of RNA from PLMVd-infected peach leaves. The RT-LAMP products generated by these experiments were analyzed using electrophoresis on 2% agarose gels.
Results
Primer Efficiency
Multiple sequence alignment revealed conserved sequences in the PLMVd genome, and six sets of PLMVd-LAMP primers were designed on the basis of the sequence LC469622 (Table 1). The primer sets were evaluated using total RNA from each of the peach pollens, and primer efficiency was determined using both SYBR Green I staining and agarose gel electrophoresis. After testing the efficiency of six sets of primers, only one set of primers (PLMVd-6) was selected for the RT-LAMP assay for PLMVd detection. Amplification was only achieved by one of the primer sets (PLMVd-6; Fig. 1), so the PLMVd-6 primer set was used for all further assays.

Fig. 1
Effectiveness of RT-LAMP primer sets for detecting peach latent mosaic viroid. (A) Visualization of RT-LAMP products using agarose gel electrophoresis. (B) Visualization of RT-LAMP products using SYBR Green I (orange: negative; yellow: positive). Lane M: 100-bp DNA ladder; P1–P6: primer sets; lane P: viroid-infected peach pollen; lane N: non-template control.
RT-LAMP Optimization
Total peach pollen RNA was used to determine the optimal temperature and reaction time for detecting PLMVd (Figs. 2 and 3). At 60°C, 63°C, and 66°C, the 60-min incubation yielded relatively high amplicon concentrations, and the products produced both visual color changes (SYBR Green I) and ladder-like patterns (Fig. 2). Based on band shape and intensity, 63°C was selected as the optimal reaction temperature. At this temperature, no amplification was observed between 5 and 20 min, and only weak amplification was observed at 30 min. Incubation at 63°C for 60 and 70 min yielded the clearest and highest intensity electrophoresis bands. Therefore, all additional RT-LAMP assays were performed using incubation at 63°C for 60 min (Figs. 2 and 3).
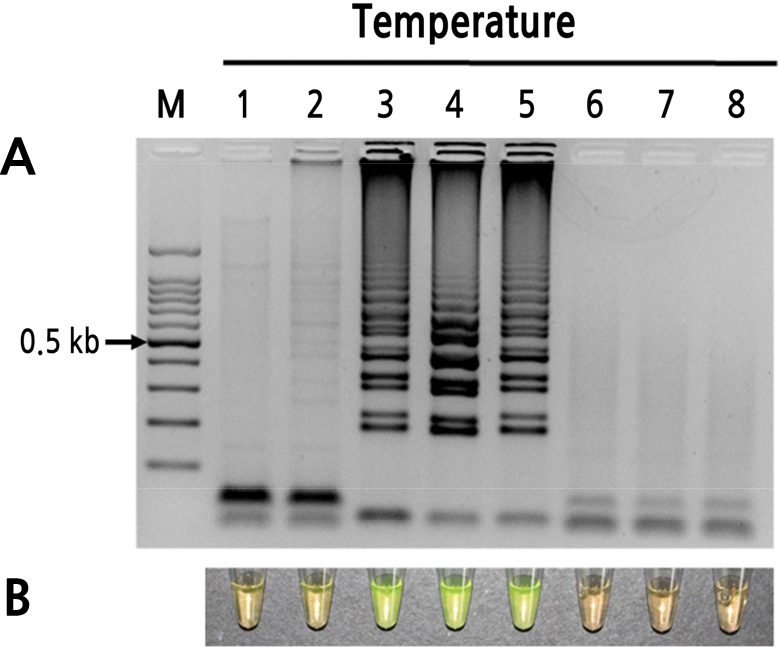
Fig. 2
Optimum reaction temperature of the RT-LAMP assay for detection of PLMVd. (A) Visualization of RT-LAMP products using agarose gel electrophoresis. (B) Visualization of RT-LAMP products using SYBR Green I (orange: negative; yellow: positive). Lane M: 100-bp DNA ladder; lanes 1–8: 54°C, 57°C, 60°C, 63°C, 66°C, 69°C, 72°C, and 75°C, respectively.
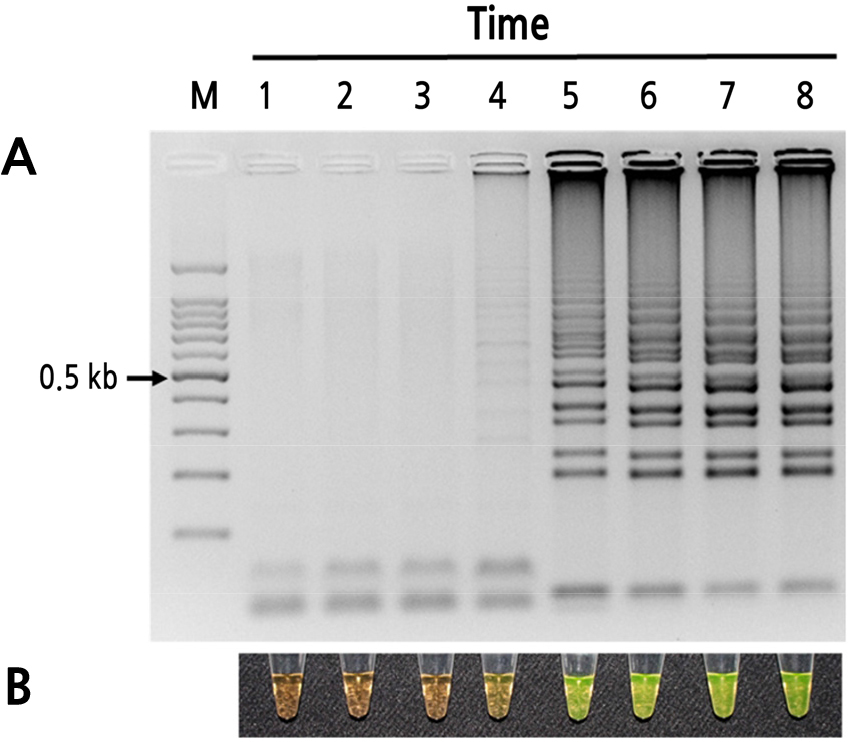
Fig. 3
Optimum reaction time of the RT-LAMP assay for detection of PLMVd. (A) Visualization of RT-LAMP products using agarose gel electrophoresis. (B) Visualization of RT-LAMP products using SYBR Green I (orange: negative; yellow: positive). Lane M: 100-bp DNA ladder; lanes 1–8: 5, 10, 20, 30, 40, 50, 60, and 70 min, respectively.
Sensitivity and Specificity of RT-LAMP for PLMVd Detection
To evaluate the sensitivities of the assays, RT-LAMP and RT-PCR were performed using a dilution series of PLMVd RNA (10-fold serial dilutions), with concentrations ranging from 1.3 × 10-1 to 1.3× 10-8 µg per reaction. RT-PCR yielded only weak amplification of 1.3 × 10-4 µg template, whereas RT-LAMP yielded much greater amplification, as indicated by agarose gel analysis (Fig. 4). Thus, RT-LAMP was approximately 10-fold more sensitive than conventional RT-PCR for the detection of PLMVd. The specificity of RT-LAMP for PLMVd was performed using total RNA extracted from peach leaves infected by ACLSV or PNRSV, which are highly destructive to peach trees in Korea. The samples were also tested for the presence of ACLSV and PNRSV using RT-PCR with specific primers. Using optimal reaction temperature and time, only the PLMVd-infected samples yielded amplification, and no amplification was observed for samples infected with ACLSV or PNRSV (Fig. 5). Thus, the RT-LAMP assay was highly specific for PLMVd.

Fig. 4
Relative sensitivity of RT-LAMP and RT-PCR for detecting peach latent mosaic viroid. (A) Visualization of RT-LAMP products using agarose gel electrophoresis. (B) Visualization of RT-LAMP products using SYBR Green I (orange: negative; yellow: positive). (C) Visualization of RT-PCR products. Lane M: 100-bp DNA ladder; lanes 1–8: 10-1, 10-2, 10-3, 10-4, 10-5, 10-6, 10-7, and 10-8 dilutions of viroid RNA, respectively.
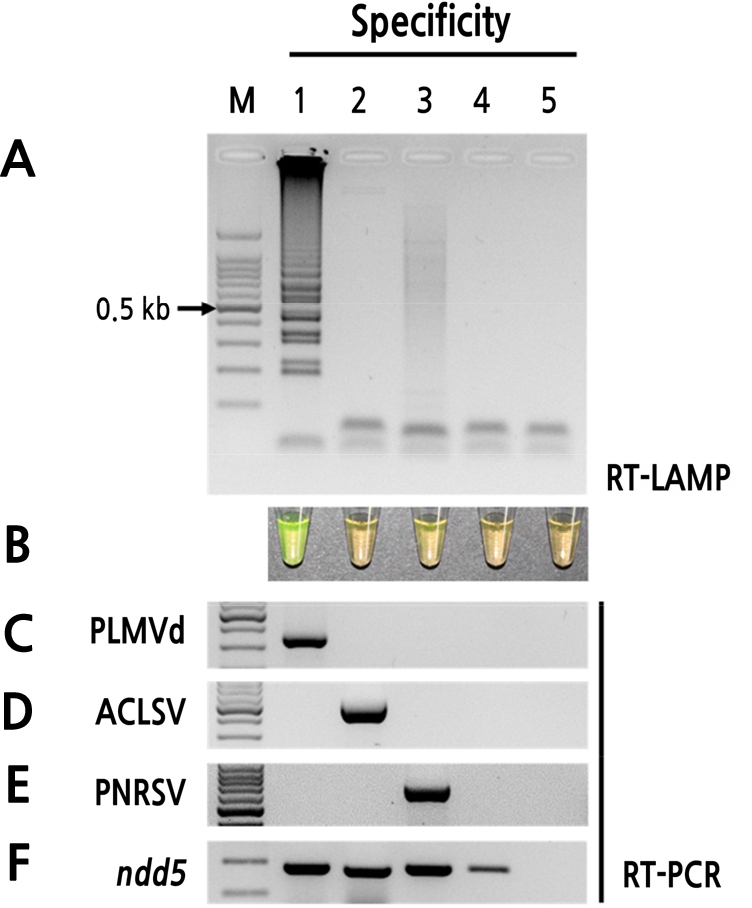
Fig. 5
Specificity of RT-LAMP assay for detection of PLMVd. (A) Visualization of RT-LAMP products using agarose gel electrophoresis. (B) Visualization of RT-LAMP products using SYBR Green I (orange: negative; yellow: positive). (C) RT-PCR amplification of PLMVd. (D) RT-PCR amplification of apple chlorotic leaf spot virus (ACLSV). (E) RT-PCR amplification of prunus necrotic ring spot virus (PNRSV). (F) RT-PCR amplification of nad5 (internal control). Lane M: 100-bp DNA ladder; lane 1: viroid-infected peach pollen; lane 2: ACLSV-infected peach pollen; lane 3: PNRSV-infected peach pollen; lane 4: healthy peach pollen (negative control); lane 5: non-template control.
Detection of PLMVd in Field Samples
To test the reliability of the RT-LAMP assays for detecting PLMVd, six peach pollens that were imported from China in 2018 and 2019 and two peach leaves that were randomly collected from the field were tested. PLMVd was detected in the peach pollen and leaves by both RT-LAMP and RT-PCR (Fig. 6). Thus, the RT-LAMP assay is an effective and reliable method for detecting PLMVd in field-collected samples.
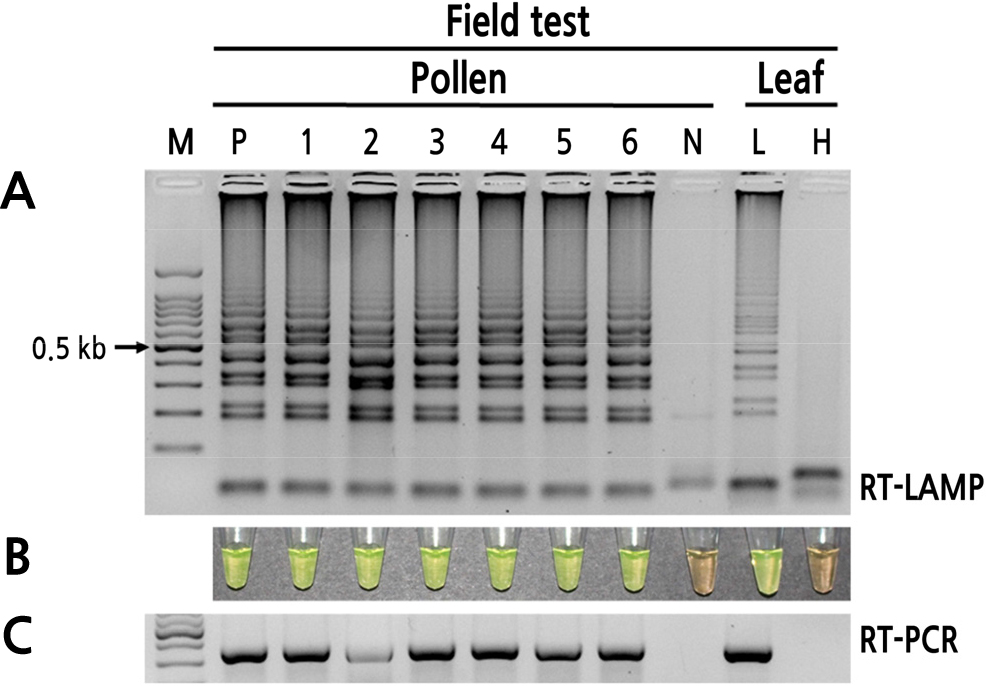
Fig. 6
Field test of RT-LAMP for PLMVd detection. (A) Visualization of RT-LAMP products using agarose gel electrophoresis. (B) Visualization of RT-LAMP products using SYBR Green I (orange: negative; yellow: positive). (C) Visualization of RT-PCR products. Lane M: 100-bp DNA ladder; lane P: viroid-infected peach pollen; lanes 1–3: peach pollens imported in 2018; lanes 4–6: peach pollens imported in 2019; lane N: healthy peach pollen (negative control); lane L: PLMVd-infected peach leaf; lane H: healthy peach leaf (negative control).
Discussion
PLMVd causes serious economic losses in the cultivation of several important fruit trees, including apricot, cherry, pear, nectarine, plum, peach, and apple (Dubé et al., 2009). PLMVd is globally distributed and can be transmitted by grafts, aphids (Myzus persicae), and contaminated hand tools, as well as by pollen (Barba et al., 2007). Therefore, rapid and reliable detection methods are needed to exclude the introduction of high-risk pathogens via fruit tree pollens.
Both RT-PCR (Jo et al., 2016; Li et al., 2012; Shamloul et al., 1994) and qRT-PCR (Luigi and Faggioli, 2011; Parisi et al., 2011) are often used to detect PLMVd. However, RT-PCR-based techniques are time-consuming, have high background and low specificity, and require laborious post-amplification analysis or costly equipment. In contrast, the one-step RT-LAMP assay resolves these issues, and field diagnostics can be further simplified using SYBR Green I, which allows amplification to be confirmed using direct visual observation (Notomi et al., 2000) without the need for electrophoresis.
The aim of the present study was to develop a rapid, sensitive, and reliable method for detecting PLMVd in peach pollen. One-step RT-LAMP could be performed in a single tube by mixing the AMV reverse transcriptase into the LAMP reaction mixture since the AMV reverse transcriptase was not negatively affected by the assay’s high temperature (Fukuta et al., 2003). In the present study, RT-LAMP was able to amplify PLMVd from pollen and leaf samples in 40 min using a water bath and SYBR Green I. Because the six RT-LAMP primer sets were designed using conserved regions of the PLMVd genome (data not shown), the assay is suitable for detecting PLMVd isolates worldwide. The optimal RT-LAMP conditions for detecting PLMVd were 63°C and 60 min, whereas conventional RT-PCR required at least 2–3 h to detect PLMVd. Furthermore, the RT-LAMP method was highly specific for PLMVd and was approximately 10 times more sensitive than RT-PCR (Fig. 7). Therefore, RT-LAMP appeared to be suitable for the rapid detection of PLMVd in field samples of either peach pollen or leaves.
In conclusion, the present study reports an optimized RT-LAMP assay for the rapid and specific detection of PLMVd in peach pollen and leaves. In terms of equipment, the assay requires only a heated water bath and thus can be easily applied under simple field conditions, even in developing countries. Considering its simplicity, speed, and cost-effectiveness, the technique could easily be applied to the large-scale detection of PLMVd in field conditions, sanitary certification programs, and quarantine inspections.