Introduction
Materials and Methods
Plant Materials and Heat Treatment
RNA Isolation and Quantitative PCR
Transcriptome Analysis and Selection of DEG
Results
Analysis of the Transcriptome in Grapevines Exposed to High Temperature
Analysis of the Expression of the Selected DEG in Response to High Temperature in Grapevine
Discussion
Introduction
Grapevines (Vitis spp.) are an economically important fruit crop cultivated worldwide, including in Korea. Grapes are consumed as fresh fruit as well as used for making wines, juices, grape jam, and grape seed oil (Park et al., 2010; Salazar-Parra et al., 2012). Recently, rapid climate change has resulted in serious issues such as decreased production and deterioration in quality (Szenteleki et al., 2012; Son et al., 2014). Grapevine genes reveal a different expression pattern in response to high temperatures (Liu et al., 2012). Recent studies analyzing the response against high temperature focused on grape berry development and ripening (Cramer et al., 2007; Deluc et al., 2007). Grapevines maintained at over 40°C for several days show poorer quality of fruit, drastically reduced efficiency of photosynthesis, and inhibited accumulation of sugar and anthocyanins (Cohen et al., 2012; Greer and Weedon, 2013).
While cool conditions are favorable for berry skin coloration, a continuous high temperature generally inhibits the accumulation of pigments such as anthocyanins due to changes in physiological activities, and expression of genes related to metabolism in the skin of grape berries (Kliewer and Torres, 1972; Mori et al., 2005; Matsumoto et al., 2007; Mori et al., 2007; Kim et al., 2015).
The expression of numerous genes is modified in grapevines under high temperature due to the activation of various mechanisms including the expression of defense related genes, metabolism-related genes, and transcription factors (Liu et al., 2012). Temperature affects many photosynthetic and molecular processes, and temperatures above 40°C are reported to reduce photosynthesis in most plants (Saxe et al., 2001; Bita and Gerats, 2013). Stress caused by high temperatures on leaves has a direct impact on the quality of grape berries (Guidoni et al., 1997; Koyama et al., 2012; Zha et al., 2016).
Understanding the molecular mechanisms of berries and vine leaves in response to high temperature is important for developing novel cultivars of grapevines tolerant to temperature changes, as well as to benefit grape production by regulating the expression of genes related to stable berry development regardless of climatic changes. It is therefore necessary to elucidate the mechanism of complex responses in leaves of grapevines exposed to high temperature in order to help maintain stable berry development and good ripening. This study was undertaken to analyze the transcriptome, and to select differentially expressed genes (DEG) and validate their expression in leaves of ‘Kyoho’ grapevines exposed to high temperature.
Materials and Methods
Plant Materials and Heat Treatment
For transcriptome analysis, leaves of grapevine (Vitis labruscana) cultivar ‘Kyoho’ were collected from vines grown in a temperature gradient greenhouse in the Agricultural Research Center for Climate Change, Jeju, Korea. The temperature gradient greenhouse was divided into T1 (low: 25°C), T2 (medium: 30°C), and T3 (high: 35°C) zones, and the temperature difference between the inlet and outlet was 4°C to 5°C. The grapevines were placed in a row from the inlet to the outlet and the leaves were sampled at the highest and lowest temperature, respectively.
For quantitative PCR analysis, shoots of ‘Kyoho’ grapevines from rooted cuttings with 15-20 leaves were grown in an experimental greenhouse of Yeungnam University, Gyeongsan, Korea at 25°C/18°C (day/night) and 65% relative humidity. Grapevines were kept in growth chambers (Ilshin Tech., Daejeon, Korea) equipped with a temperature controller and incubated at 25°C and 35°C for 24 h. Each treatment was conducted with three plants. Three to five leaves were randomly collected at 24 h after the treatment. All leaves sampled for RNA extraction were quickly frozen in liquid nitrogen and stored at -80°C until analysis.
RNA Isolation and Quantitative PCR
RNA isolation from ‘Kyoho’ leaves from three biological replicates was performed using a pine tree method described by Chang et al. (1993) with a slight modification in centrifugation (3,200 rpm for 30 min). RNA quality and yield were determined based on the absorbance at 230, 260, and 280 nm, which was measured using a UV/Vis Nano Spectrophotometer (MicroDigital Nabi, GyungGi, Korea).
cDNA was synthesized from RNA (500 ng) using the GoScript Reverse Transcription System (Promega, Madison, WI, USA), and quantitative PCR was performed with SYBR Premix Ex (TaKaRa Bio Inc., Osaka, Japan). The reaction conditions were conducted by subjecting the samples to one cycle at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. The top 10 genes with induced and suppressed expression were selected, and their expression levels were confirmed by reverse transcription quantitative PCR.
Transcriptome Analysis and Selection of DEG
High-quality RNA samples were prepared using the TruSeq SBS v5 protocol, after which the libraries were sequenced using an Illumina HiSeq 2000 (Illumina, San Diego, CA, USA) for paired-end reads. De novo assembly of trimmed reads was carried out using Velvet (Version 1.2.07) and Oases (Version 0.2.08), with k-mer sizes of 61, 65, 67, and 69, a minimum contig length of 200 bp, and an insertion length of 300 bp. The longest transcripts from each locus (k = 69) were selected for assembly based on the putative amino acids obtained from nucleotide sequences. Gene expression was evaluated using the short reads number, differentially expressed genes (DEGs), clustering, and annotation. DEGs due to exposure to high temperature were selected by the DESeq Bioconductor package (version 1.6.0) (Anders and Huber, 2010) with a false discovery rate (FDR) of the Benjamini-Hochberg multiple tests of 1% (p < 0.01) based on read counts (more than 500) and a minimum fold change of 1.5 in at least one pairwise comparison. Gene expression patterns were determined using Pearson’s correlation of hierarchical clustering with at least a two-fold change in expression between the control and treated plants. Genes expressed in the high-temperature treatment were annotated using BLASTN and BLASTX in combination with information from the gene ontology (GO) (Ashburner et al., 2000) and KEGG pathway (Kanehisa et al., 2004). The information retrieved by GO analysis was divided into biological process (BP), cellular component (CC), and molecular function (MF), according to the relevant gene function. The selected DEG were spotted on the KEGG pathway through GO analysis.
Results
Analysis of the Transcriptome in Grapevines Exposed to High Temperature
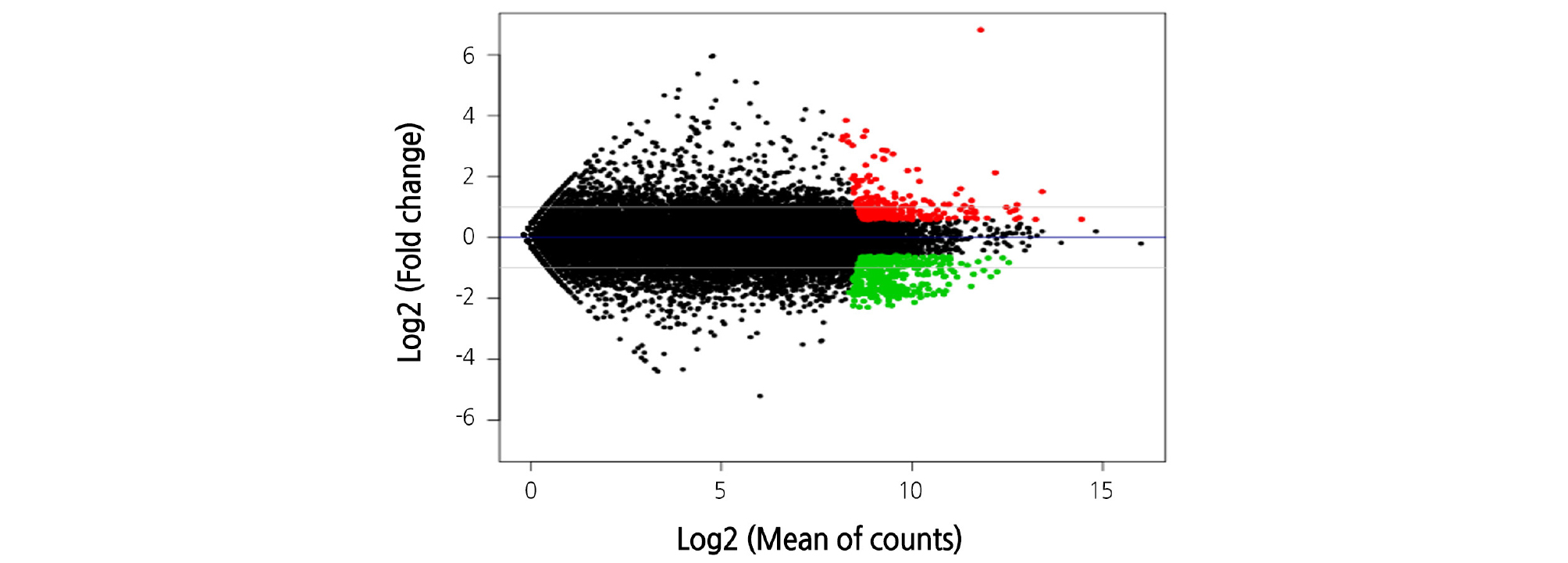
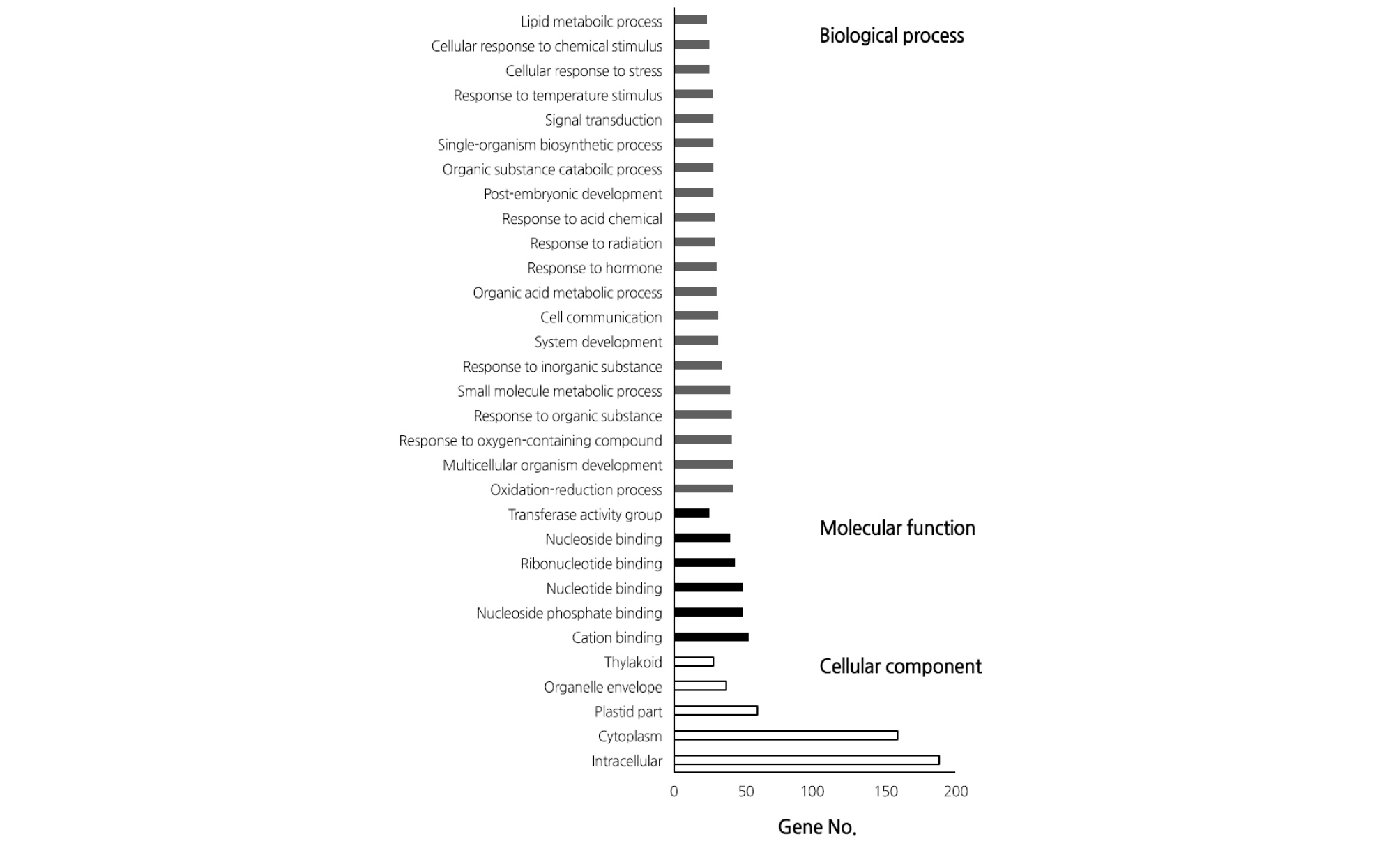
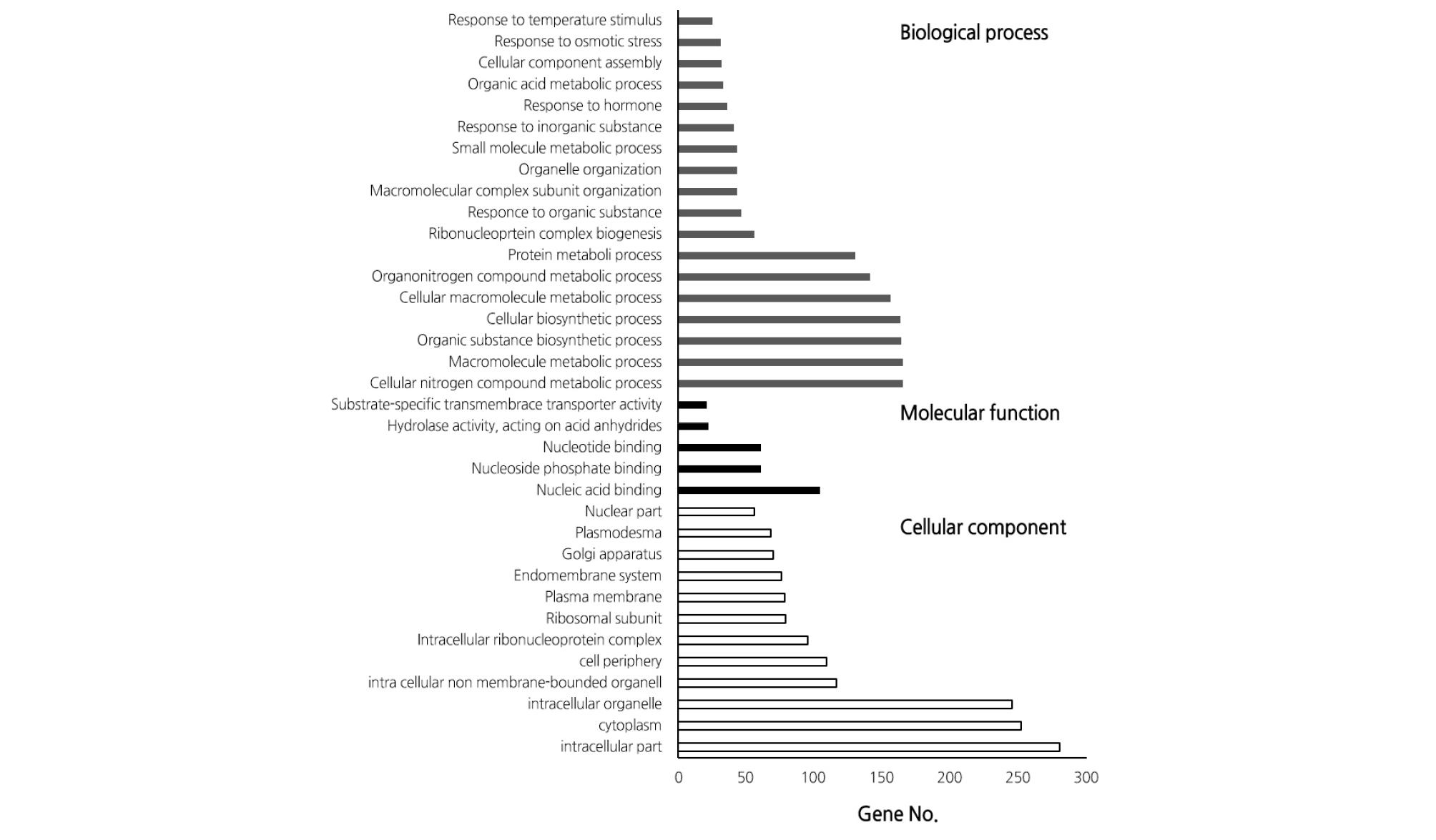
A total of 92 million transcripts were used for transcriptome analysis with a total length of 7 billion bp and an average read length of 89.6 bp; an average of 85.5% of the data were used for further analysis (Table 1). As a result of selection of the DEGs in response to high temperature, we identified 243 up-regulated genes and 378 inhibited genes (Fig. 1 and Table 2). Classification of the DEGs with induced expression in GO term level 3 showed that 49 genes were involved in metabolism, 22 genes in cell structure, and 15 genes in molecular function (Fig. 2). In contrast, DEGs with inhibited expression in GO term level 3 revealed the involvement of 34 genes in metabolism, 32 genes in cell structure, and 16 genes in molecular function (Fig. 3).
Table 1. Statistical data from the transcriptome analysis of grapevine leaves exposed to high temperature
Table 2. Data of genes showing differential expression patterns at the transcriptome level in grapevine leaves exposed to high temperature
| Control vs. treatment | Regulation pattern | 2-Fold change | 1.5-Fold change | |||
| No. of DEGs | No. of annotated DEGs | No. of DEGs | No. of annotated DEGs | |||
| Leaf 25°C:35°C | Up | 32 | 30 | 252 | 243 | |
| Down | 95 | 92 | 391 | 378 | ||
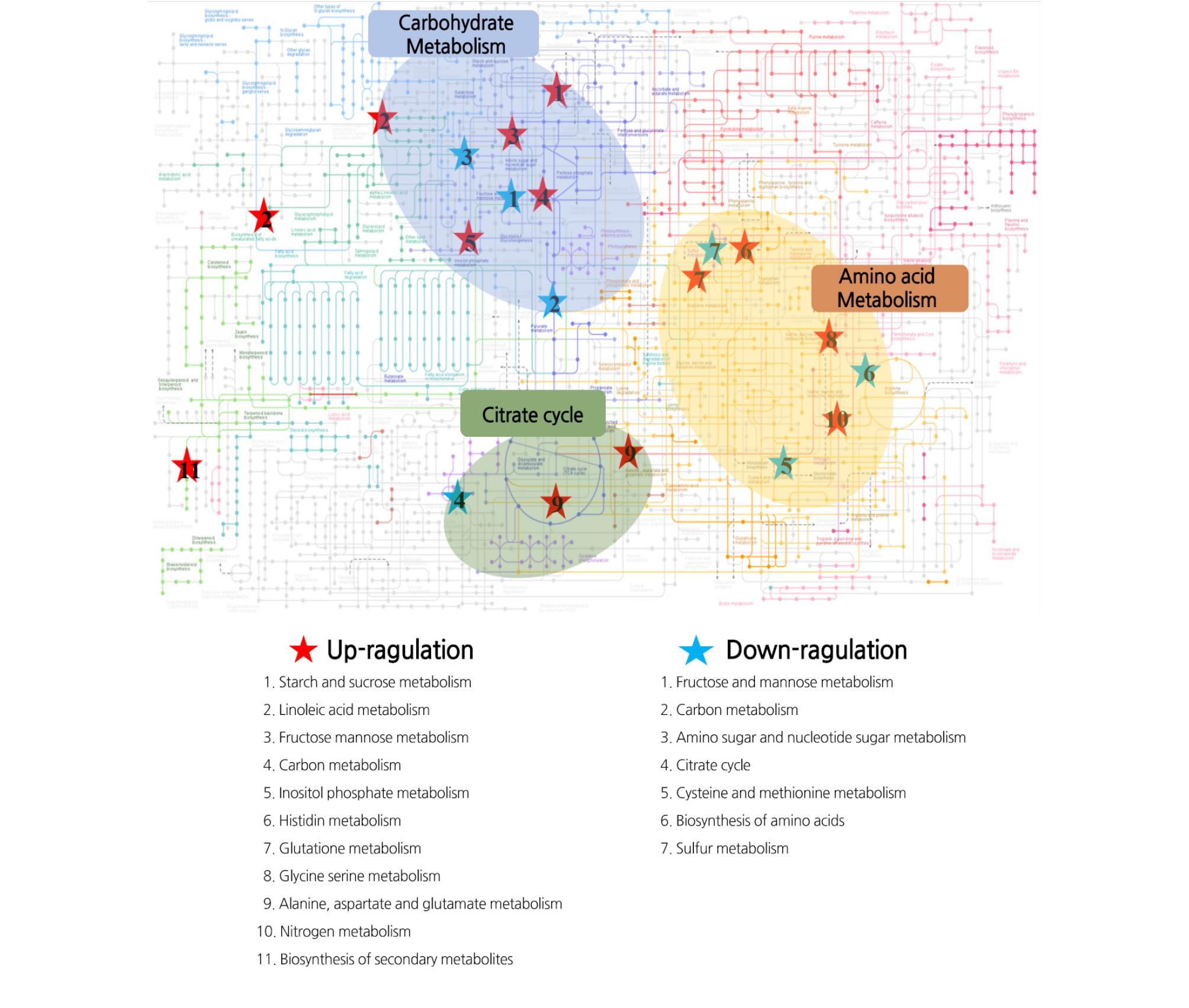
Functional classification of DEGs by GO analysis revealed that most of the induced DEGs were involved in the metabolism of starch and sucrose, linoleic acid, fructose, mannose, carbon, inositol phosphate, histidine, glutathione, glycine, serine, alanine glutamate, and nitrogen, and biosynthesis of secondary metabolism. On the other hand, inhibited DEGs were involved in the metabolism of fructose and mannose, carbon, amino sugars and nucleotide sugars, citrate cycle, cysteine, and methionine, and biosynthesis of amino acids, and sulfur (Fig. 4). These metabolism pathways were mainly clustered with carbohydrate metabolism, amino acid metabolism, and citrate cycle in the KEGG metabolic pathway (Fig. 4).
Analysis of the Expression of the Selected DEG in Response to High Temperature in Grapevine
Among the selected DEGs induced by high temperature, the top 10 genes with induced expression include BCL-2- associated athanogene 6, heat shock protein 21, Haloacid Dehalogenase (HAD) superfamily, and subfamily IIIB acidphosphatase, whereas the top 10 genes with inhibited expression include APS reductase3, ribosomal protein S7e family protein, and ribosomal protein L2 family (Table 3).
Table 3. The top 10 genes highly up-regulated (U) and down-regulated (D) in grapevine leaves exposed to high temperature
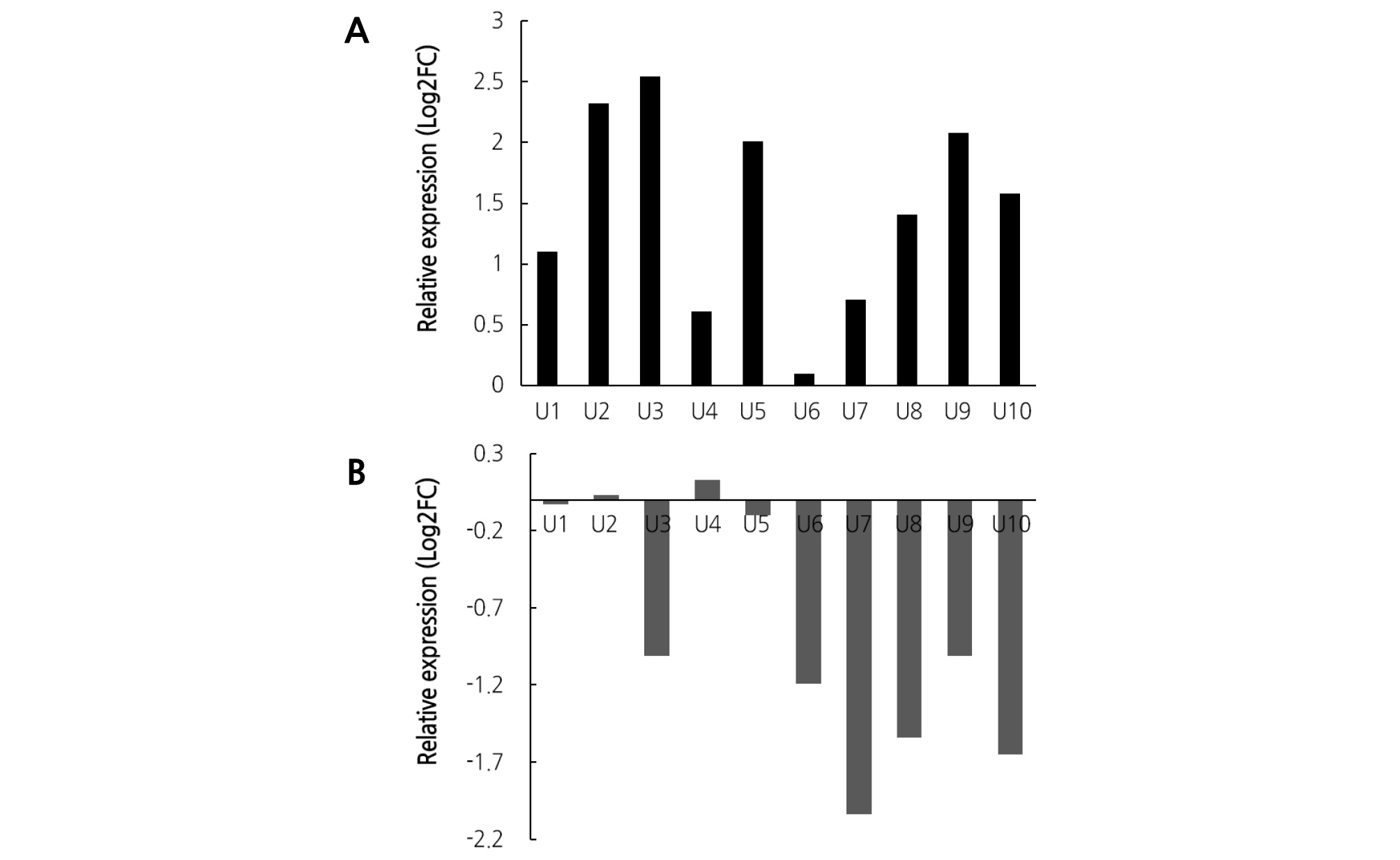
The selected genes were evaluated for their differential expression levels by reverse transcription followed by quantitative PCR (RT-qPCR) using the primer pairs listed in Table 4, which were designed based on the transcript sequences. The results of RT-qPCR analysis using gene-specific primers revealed that the top 10 up-regulated and top 10 down-regulated genes were differentially expressed, and their expression levels were similar to those determined from the transcriptome analysis (Fig. 5).
Table 4. Primer list and nucleotide sequences used for real-time PCR analysis
Discussion
High temperatures are a major issue in crop production, resulting in reduced yields and poor quality of berries in grape production (Zhang et al., 2005). Temperature is the most important environmental factor in viticulture, and grapevine activates various genes related in metabolic processes including carbohydrate metabolism and amino acid metabolism during bud and vine growth and berry development of grapes in optimal temperature condition (Kaplan et al., 2007; Kim et al., 2016, Kim et al., 2017). A transcriptome analysis of grape berries exposed to high temperature reported that expression of genes, including isocitratelyase, cysteine proteinases superfamily protein, and cupin family proteins, were highly induced, and expression of genes including flavanon3-hydroxylase, phenylalanine (PHE) ammonia lyase 1, and chlorophyll A-B binding family protein were the most inhibited (Kim et al., 2015; Kim et al., 2018). It is also reported that sulfur metabolism plays an important role in the stress response in grapevines (Koprivova and Kopriva, 2008)
The BCL-2-associated athanogene 6 gene, the most abundant transcript in grapevine leaves exposed to high temperature, is related to apoptosis and is known to be induced by various stimuli. BCL-2-associated athanogene 6 was also found to be highly expressed in grape berries exposed to high temperature (Kim et al., 2018). Apart from high temperature, the BCL-2-associated athanogene 6 gene was also expressed in response to hormones, low temperature, salt, drought, and oxidative stress (Nawkar et al., 2017). In this study, we found that among the DEGs, the BCL-2-associated athanogene 6 gene is induced after exposure to high temperature in the leaves of grapevines, which seems to be closely related to the response to high temperature in grapes.
Heat shock protein (HSP) genes were also induced by high temperature in this study. The main HSPs include HSP60, HSP70, HSP90, HSP100, and small HSPs, which are classified depending on their structure (Huang et al., 2008). It was reported that expression of various HSPs was regulated in grapevine leaves after exposure to high or low temperature (Liu et al., 2012; Kim et al., 2016, Kim et al., 2017). In the current study, the HSP gene detected abundantly in the transcriptome analysis is considered to be related to its activation in response to stresses caused by exposure to high temperature.
The expression of abscisic acid (ABA)-induced phosphatase 2C (PP2C) gene 3 is differentially regulated by high and low temperatures, as well as by drought and ABA stress (Timucin and Sezerman, 2018; Yang et al., 2018). Consist with these reports, we observed that the ABA-induced PP2C gene 3 is also highly induced in grapevines exposed to high temperature.
OBF‐binding protein 3 (OBP 3) responsive gene 1 is reported to be strongly induced by iron (Fe) deficiency in Arabidopsis thaliana roots (Buckhout et al., 2009); this gene is also related with the response to high temperature in grapevines.
In the group of genes whose expressions are inhibited by high temperature, adenosine-5'-phosphosulfate (APS) reductase 3 is reported to play an important role in the salt stress response (Koprivova and Kopriva, 2008). The expression of tonoplast intrinsic protein 1 in wheat (Triticum sp.) leaves was strongly inhibited in the 4 h drought stress treatment. In contrast, expression of tonoplast intrinsic protein 1 was induced in the 8 h drought stress treatment (Cevher-Keskin et al., 2015). Another study reported that Ferritin 2 in grapevines plays an important role in the stress response (Kós et al., 2008).
The pathogenesis-related gene 1 (PR-1) reportedly enhances the resistance to biological and non-biological stresses (Ali et al., 2018). Induced expression of the tonoplast intrinsic protein1 gene has been reported under stress conditions of nitrogen deficiency in Arabidopsis (Loqué et al., 2005). However, in the current study, we observed inhibited expression of PR-1 and the tonoplast intrinsic protein1 gene in response to high temperature, which indicates that further investigation is needed regarding their role in response to high temperature in grapevines. Contrarily, Zha et al. (2016) reported that a 45°C treatment showed no irreversible effect on the growth of leaves under field conditions; the growth and development of grapes remained constant, and was stimulated in response to heat. The current study demonstrates that most of the genes whose expression was altered by high temperature are involved in the mechanism of response to stress in plants.
Taken together, our results suggest that there is a possible relationship between the genes expressed at high temperature and defense responses against stress in grapevine leaves. Moreover, information about changes in expression of various genes at high temperatures will be a valuable resource for breeding grapevines tolerant to high temperatures. In addition, genes that are specifically expressed by high temperatures can be used as molecular markers for selection of heat resistant grape varieties, and help elucidate the defense mechanisms in fruit trees against high temperature.